Would you like some Hi-C with that Woolly Mammoth?
Woolly Mammoths were just big, furry, elephants, right? Maybe, and now we have the tools to compare the structures of their genomes.
The Woolly mammoth is the best studied of the mammoths because their remains have been found frozen and incredibly well preserved in Siberian and North American permafrost.
This has allowed for the collection of tissues from these ancient creatures who roamed the earth over 800,000 years ago.
Through genetic analyses, it has been shown that the closest living relative of the Woolly Mammoth is the smaller (and noticeably less woolly!) Asian elephant.
But performing genetic analyses on ancient specimens is tricky, even if they are well preserved in ice for millennia!
The half-life of DNA is ~ 500 years, and even if totally frozen, it’s estimated that every bond in a DNA molecule would be broken after about 7 million years.
So, suffice it to say, ancient DNA is highly fragmented, which makes it tough to reconstruct genomes of ancient organisms!
In many cases, paleontologists use a close living relative as a reference for genetic studies, but as we’ve seen in humans, this can create issues with properly aligning sequences to the appropriate locations in a genome.
And much like in humans, we’ve also come to appreciate the importance of epigenetics, chromatin structure and gene regulation in helping to define the differences that we see in an organism’s appearance.
But if ancient samples are already fragmented, how can we ever get structural information out of them?
With Hi-C.
This is a molecular technique that allows you to identify sequences of DNA that are physically close to one another.
It’s also perfect for reconstructing genomic maps and identifying what parts of the genome are actively expressing genes (because DNA that touches is usually open!)
Could it be possible for us to figure out what put the wool on the Woolly Mammoth???
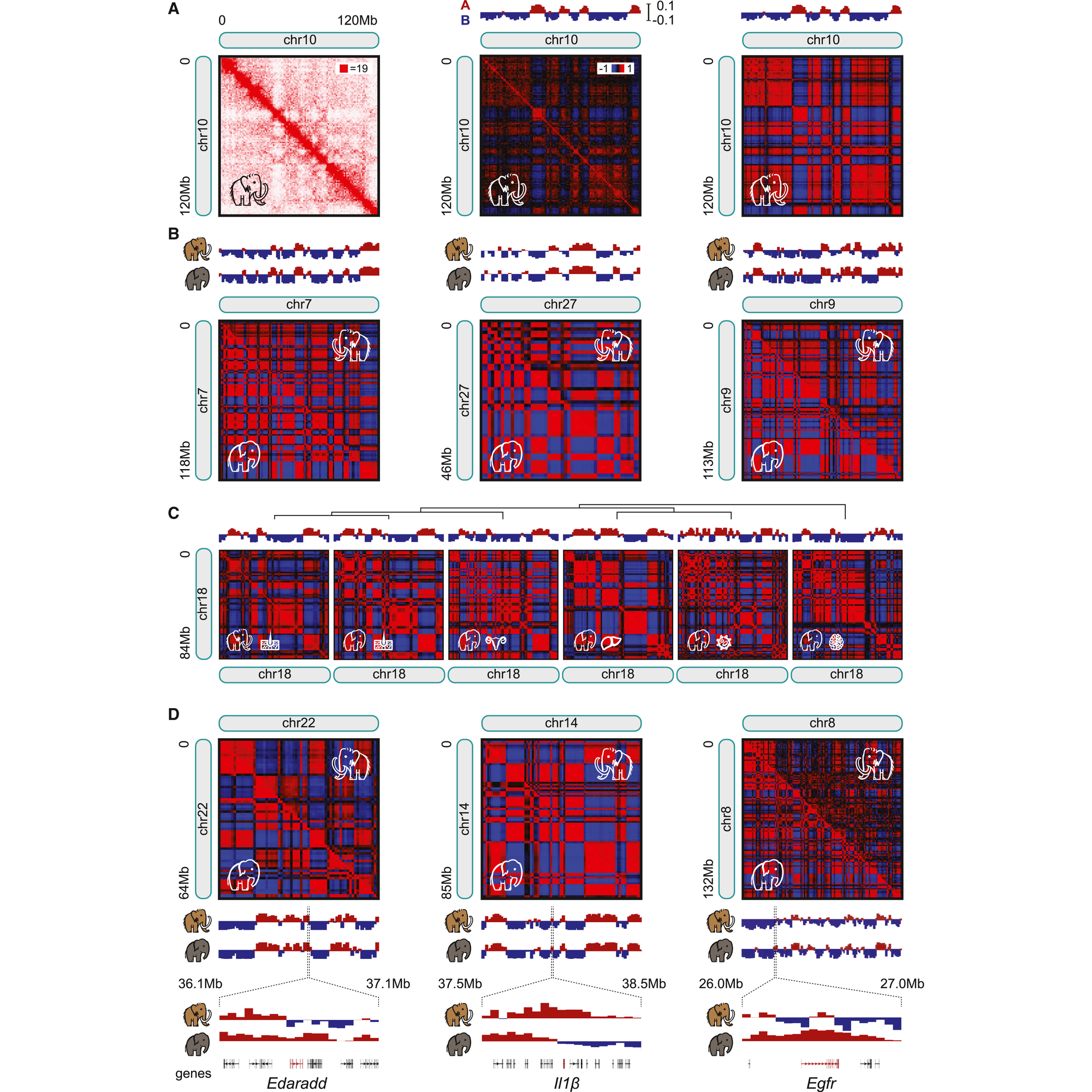
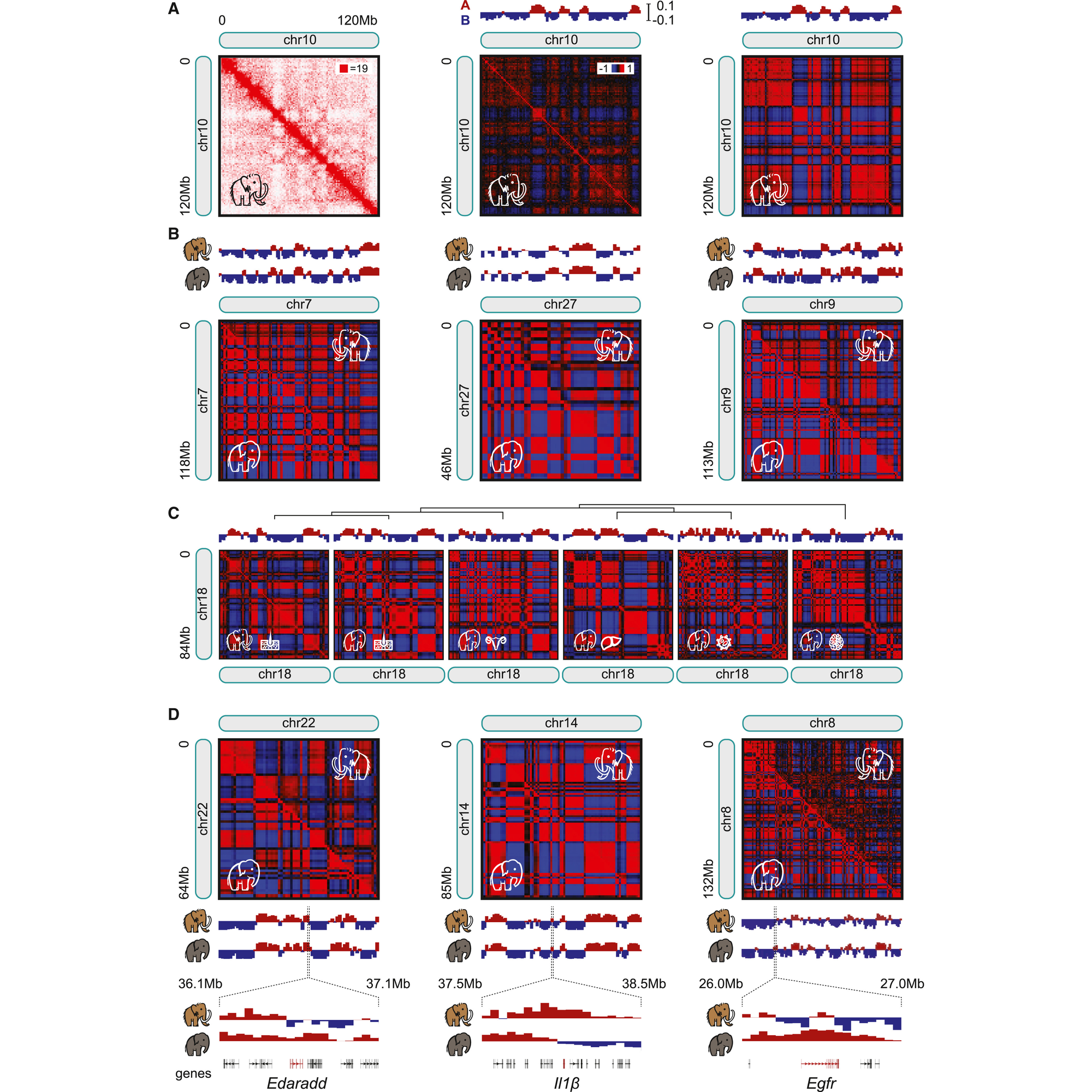
Maybe, but let’s not get ahead of ourselves, the researchers behind this paper first needed to show that it was possible to perform Hi-C on preserved mammoth samples and that they could get reasonable data out of them.
That experiment can be seen in the figure above where in a) they generate contact maps for chromosome 10, b) compare maps for chr7, 27, and 9 between mammoths and Asian elephants noting high correlation between the two animals, c) show that maps for skin are highly correlated but that chromosome architecture differs in other tissues (meaning, they’re getting real signal from mammoth skin!), d) there are sub-chromosomal differences in chromatin structure when looking at specific genes
So, why should you care?
This paper shows that techniques like Hi-C can be used to help us better understand the critical genetic (and epigenetic!) differences that define how organisms have evolved over time.
###
Sandoval-Valesco M, et al. 2024. Three-dimensional genome architecture persists in a 52,000-year-old woolly mammoth skin sample. Cell. DOI: 10.1016/j.cell.2024.06.002