Watch out, Structural Variants are coming to MRD testing!
SNPs better watch their back, a new variant class is looking to steal the MRD limelight
The goal of minimal residual disease (MRD) testing is to determine how much disease remains in cancer patients after a solid tumor is removed or a hematologic (blood) cancer is treated.
In hematologic cancers, MRD tests using PCR were first seen in the late 1980’s.
These tests targeted the IG-TR gene rearrangements that were common in lymphoid malignancies.
PCR allowed oncologists to ditch using Southern blots to detect these changes and improvements in MRD tests led to quantitative PCR based versions in the late 90’s.
But, the challenge with using PCR for this sort of testing is you need to know what you’re looking for to detect it!
If you don’t have, or know of, a common mutation in a cancer, it’s very hard to determine the residual disease burden, or detect when a cancer comes back after treatment.
Fortunately, the advent of massively parallel sequencing in the early 2000’s gave us a new tool for tracking cancer progression and finding unique markers of disease.
But the use of high throughput sequencing as a diagnostic in oncology has been prohibitively expensive until more recently.
The advent of tumor-informed high-throughput sequencing based MRD assays really began in 2018 with Natera’s publication of their Signatera product which is a single-nucleotide polymorphism (SNP) based test.
It uses a patients’ own tumor tissue to develop a SNP fingerprint that is then assayed longitudinally (over months/years) after cancer removal to see if that DNA signature reappears in a patient’s blood sample.
Reappearance of that signal is indicative of cancer’s return, and the hope is that these MRD tests can detect this recurrence in time for us to do something about it!
But SNPs aren’t the only kinds of mutations that turn up in cancers!
They’re also full of large structural variants (SV) like gene deletions, duplications and other large genomic rearrangements.
The researchers behind today’s paper showed that these SVs can also be used to generate MRD fingerprints that perform as well as or better than their SNP based counterparts!
To do this they 15x whole genome sequenced tumor tissue from breast cancer patients to identify tumor specific structural variants and created a 16 marker fingerprint.
But instead of using that fingerprint to do MORE sequencing, they instead used an ingenious method to develop SV breakpoint specific digital PCR (dPCR) assays.
Using dPCR allows them to detect single molecules in blood without having to spend a bunch of money sequencing stuff they don’t care about!
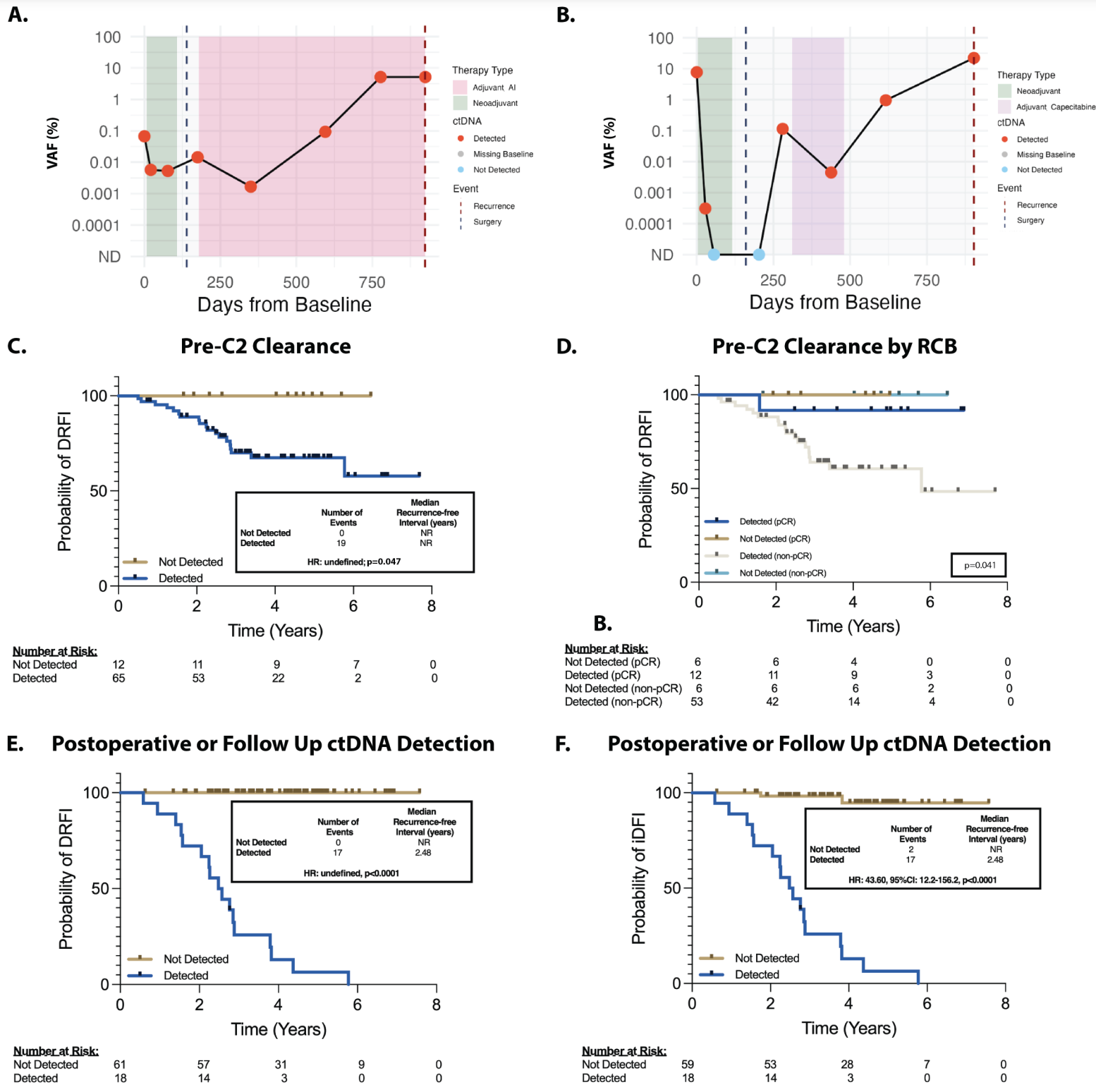
And in the figure above you can see that their hard work paid off! A and B) Show representative plots of ctDNA detection before surgery (green area), at surgery (black dashed line), after surgery and during chemo (pink and purple) and at detection of disease recurrence (red dashed line) C and D are kaplan-meier curves (track the death of subjects in a trial) showing that the detection of ctDNA in the neoadjuvant setting (before tumor was removed) was associated with poorer prognosis and E and F) that the same was true in the adjuvant setting (after the tumor was removed).
This test detected 100% of cancer recurrence with a median lead time in early breast cancer of 417 days.
That’s 417 more days to try to figure out how to delay the cancer a second time!
###
Disclosure: I have received consulting fees from SAGA Diagnostics (their Pathlight assay was used in this paper)
