Tumor immune microenvironments and how we can exploit them to fight cancer
Tumors are bad. Understanding their microenvironment is key in our effort to amp up the immune system to fight them!
Cancers are a terrible scourge on humanity.
We’ve all experienced personal losses to them, but our colloquial discussions of ‘cancer’ are overly simplistic.
‘Why haven’t we cured cancer?’
Mostly because it's not one thing.
It’s thousands, maybe millions, of things.
And treatments that are effective in even the same type of cancer aren’t effective for everyone with that type of cancer!
That’s because cancers are ‘heterogeneous’ which is a big scientific word for ‘stupidly complicated.’
And cancers are some of the most stupidly complicated diseases that we have to deal with in science.
But more recently we’ve discovered how our immune system can be used to uncomplicate things.
It’s the first line of defense when cells start misbehaving, but this defense can get exhausted and even blocked by tumor cells as they evolve mechanisms to evade detection and destruction!
One important example here is the action of the proteins PD-1 and PD-L1.
PD-1 is found on the surface of immune cells and binds to a receptor, PD-L1, that sits on the outside of non-immune cells.
This tells the immune cells to not kill them.
Mostly because you don’t want the immune system going around and killing the good guys!
Eventually, tumor cells figure out this trick too and evade the immune system by producing a bunch of PD-L1.
But because we discovered this interaction, we were able to develop one of the first cancer immunotherapies, Keytruda, to take back control of the immune system and attack these resistant cancers.
If Keytruda is around, it binds to PD-1 and blocks its interaction with PD-L1 allowing immune cells to commence the killing!
This has been a revolutionary drug to combat susceptible cancers, but what if there were other ways we could get around tumor immunity to attack cancers?
That’s where today’s paper comes in!
Because you can’t destroy tumors if you don’t know their weak points.
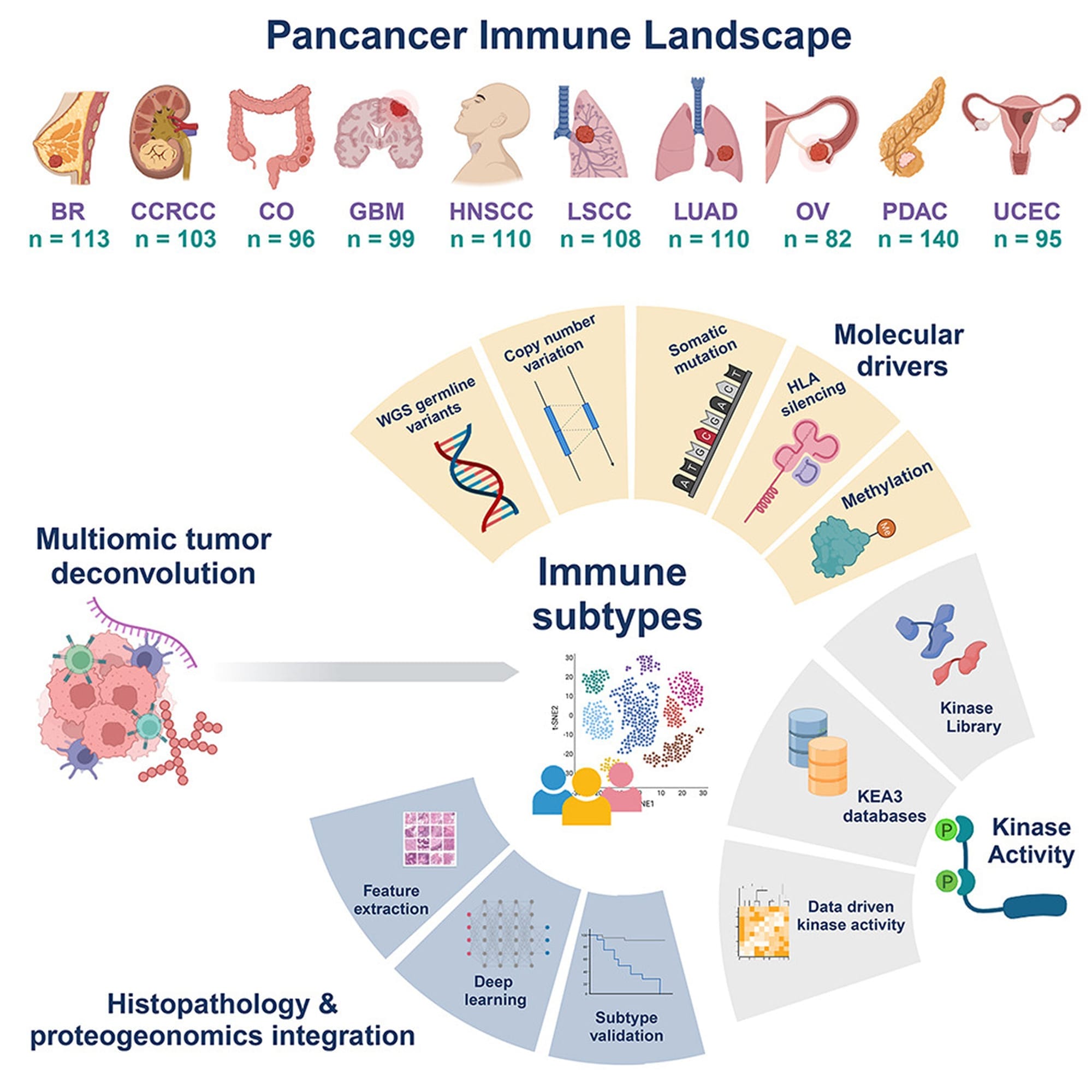
And to find those weak points, the authors cast a very wide net (see image below) looking at genetic, epigenetic, spatial transcriptomic, spatial proteomic and phosphoproteomic (kinases!) data in breast cancer (BC), renal carcinoma (CCRCC), colon cancer (CO), glioblastoma (GBM), head and neck cancer (HNSCC), two lung cancers (LSCC and LUAD), ovarian cancer (OV), pancreatic cancer (PDAC), and uterine cancer (UCEC).
There's a boat-load of information in here, and that’s not surprising since it’s a product of the Clinical Proteomic Tumor Analysis Consortium!
But what makes this dataset unique is it comprehensively profiles the tumor immune microenvironment, opening up new possibilities for targeted therapeutics based on the 'immune subtype' of each individual tumor.
