The 2-D structure of tRNA was solved in 1965, its solver probably isn't someone you've heard of before
The next great mystery to solve after the discovery of the DNA double helix was to figure out how the nucleotide sequence coded for proteins.
This was a challenging concept in the 1950's because prior to the publication of the Hershey-Chase experiment in 1952, there was a dispute over what the genetic material actually was.
Linus Pauling and other prominent scientists believed that the genetic material was protein.
They reasoned that since there were 20 amino acids, protein was obviously the genetic material because DNA only had 4 nucleotides and lacked the complexity to store information!
However, Alfred Hershey and Martha Chase showed with the help of a blender full of bacteria (and phages) that DNA, and not protein, was unequivocally the genetic material.
This presented a significant problem.
Watson and Crick get credit for solving the structure of DNA in 1953 (using data from Rosalind Franklin), but Crick's later theoretical work postulating how 4 nucleotides could code for all 20 of the amino acids was just as important.
In a lecture in 1957, he laid out 3 hypotheses (and the central dogma!):
1) RNA is the intermediary between DNA and protein
2) Certain RNAs serve as amino acid adaptors
3) These 'adaptor RNAs' use a triplet code to translate the genetic information into protein
Concurrently and without knowledge of Crick's hypotheses, Paul Zamecnik and Mahlon Hoagland discovered that small, soluble, RNAs could be charged with amino acids through an enzymatic process in 1958.
This independently confirmed Crick's adaptor hypothesis.
However, it was Robert Holley and his team at Cornell that determined the structure of this amino acid charged RNA which we now refer to as transfer RNA or tRNA.
Holley and team spent 9 years methodically sequencing tRNA using pancreatic and Taka-Diastase ribonucleases to chop up the RNAs into small fragments and then piece them back together.
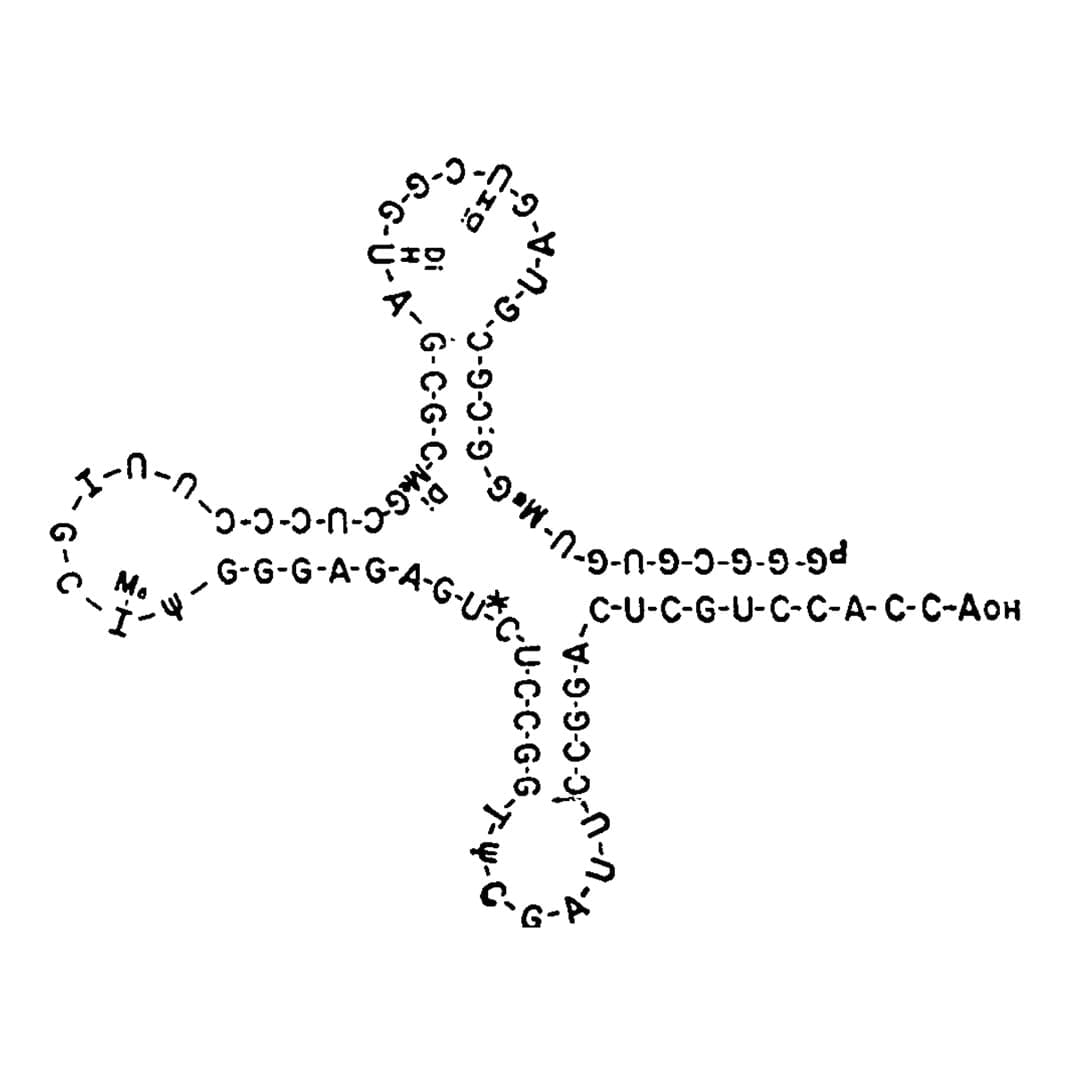
They published the complete structure of all 77 nucleotides of tRNA-ala in 1965.
Ultimately, its the structure of tRNA that explains how it functions in the translation of RNA.
Holley shared the Nobel Prize with Khorana and Nirenberg, who deciphered the triplet code, in 1968.
The Nobel winning cloverleaf structure of tRNA depicted in that 1965 publication can be seen above.
It was proposed by a member of Holley's team, Elizabeth Beach Keller.
She was acknowledged for 'helpful suggestions.'
While her name does not appear as an author nor is she credited in textbooks that contain this now famous structure, her 1997 obituary tells the story of how she modeled tRNA using pipe cleaners, paper and velcro.
She sent the final structure to Holley in a Christmas card.
Her friend, Joseph Calvo, remembered her work during that time, "Her contributions were always behind the scene, looking until she found the shape that something wanted to take.''

