The ends of our chromosomes are a paradox
What if I told you that we didn't fully understand how human DNA was copied until 1989?
Meselson and Stahl showed in 1958 that replication of the DNA double helix, the process that copies DNA, is semi-conservative.
This means that the daughter double helix contains one newly synthesized strand and one old strand that serves as the template for copying.
During the process of replication, the parent DNA helix is unwound at an 'origin of replication' and the copying process begins.
Unlike humans, and other organisms that have a linear genome, bacteria don't have to figure out how to copy the ends of their genomes!
Bacteria have a circular genome so the process finishes where it began and ends with the creation of two circles.
But, if a genome is linear, there's a big problem when the machinery hits the end of a chromosome.
You see, replication only occurs in a single direction, 5'->3'. There's a 'leading strand,' which is made contiguously as the DNA is copied, and a 'lagging strand,' which is made by creating short fragments that are connected together using an enzyme called DNA ligase.
The problem is that once the leading strand gets to the end of a chromosome, the machinery that simultaneously creates the lagging strand doesn't have enough DNA at the end to sit on to fill in that last little fragment!
The lagging strand should always end up being just a little bit shorter than the leading strand, and so, a genome should get shorter every time it's copied!
Except it doesn't.
And that's due to a repetitive sequence at the end of our chromosomes called the telomere!
But we didn't really know what the telomere did until 1975 when Elizabeth Blackburn discovered that a highly repetitive nucleotide sequence (C-C-C-C-A-A) made up the telomeres of tetrahymena (a freshwater microorganism).
Blackburn had an inkling that this repetitive sequence was important and for the next 14 years she, along with help from Jack Szostak and Carole Greider, figured out how the ends of chromosomes are copied and preserved.
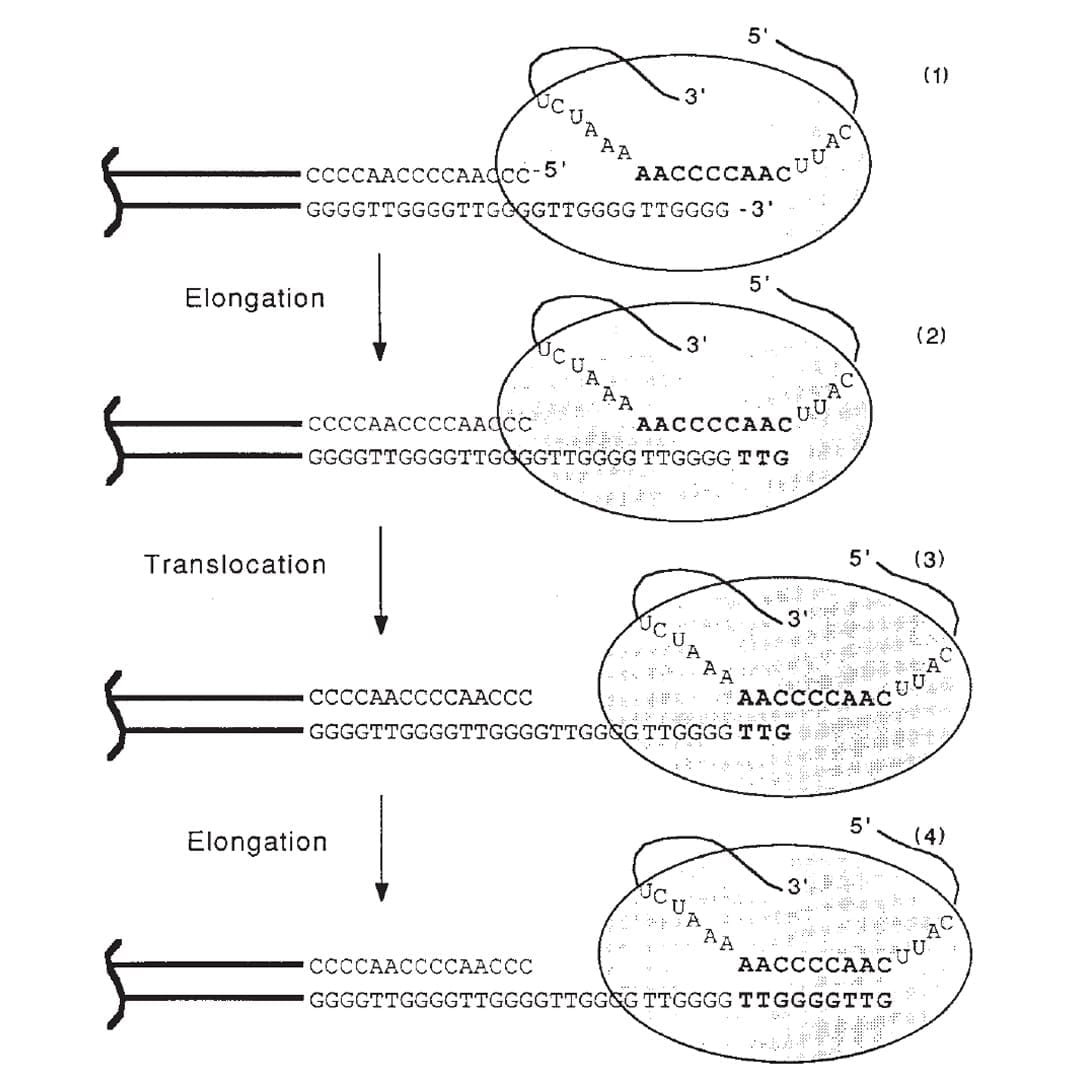
The figure above is the culmination of those years of experiments.
1) Shows the end of a chromosome and the telomerase enzyme (oval) with its RNA template bound to the TTGGGG of the telomere. Telomerase is a reverse transcriptase - these use RNA as a template to create DNA.
2) Elongation occurs, adding a TTG to the telomere.
3) Telomerase shifts down the strand (translocates) 6 bases to the new TTG.
4) Elongation occurs again.
This process of elongation and translocation is repeated multiple times, adding back sequence and preventing the shortening of the genome during replication!
Blackburn, Greider and Szostak were awarded the Nobel Prize for this discovery in 2009.

