That beating sensation you feel in your chest is the product of billions of cells working together to pump blood throughout your body.
Ever get lost in your own heart? Have no fear, there's a new atlas for that!
We now have a better idea of how those cells accomplish this!
You might not think about this very often, but your heart is critical for keeping ALL of the cells in your body alive.
It’s probably the most important organ involved in bringing nutrients to cells and removing waste from the cellular environment.
That’s not to discount the lungs, gut, liver and kidneys!
But without our blood pump, none of those organs would be able to do their jobs either.
The heart is also an incredibly complex organ composed of various structures that are essential for its rhythmic beating.
Disruption of these structures during development can lead to congenital heart disease or structural heart diseases like hypertrophic cardiomyopathies and valvulopathies later in life.
But, understanding how these diseases develop has been tough.
This is because the heart has a ton of cells that all perform slightly different functions!
These include cardiomyocytes (muscle), cardiac fibroblasts (structural support), endothelial cells (cover the inner and outer surfaces), smooth muscle cells, neuronal cells, and immune cells.
But determining the composition and interactions of these cells and how they form and maintain the crucial structures and functions of the heart has remained a challenge.
Fortunately, we now have new single-cell techniques to tease apart the answers to these questions!
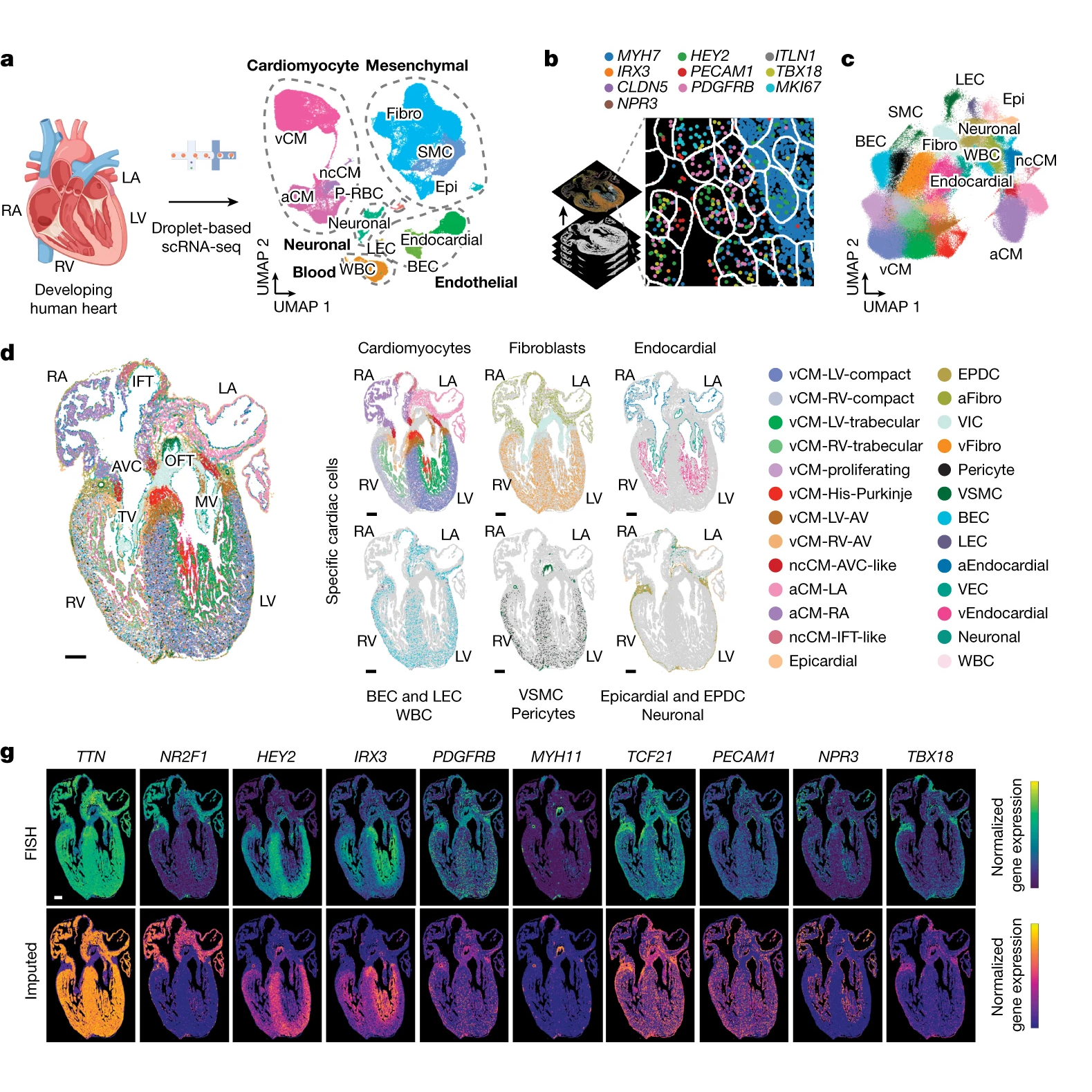
The researchers behind this week’s paper combined single-cell RNA sequencing (scRNA-seq) and multiplexed error-robust fluorescence in situ hybridization (MERFISH) on developing human hearts.
The figure above shows the major findings of the paper including an overview of the a) scRNA-seq and b and c) MERFISH techniques along with how each clustered cells through gene expression mapping.
In d) MERFISH gene expression maps were overlaid visually to show where all of the major classes of heart cells are located spatially.
And g) validates the performance of MERFISH and scRNA-seq by comparing the gene expression levels observed by each technique.
Overall, cardiomyocytes and non-cardiomyocytes showed regional and structural diversity, with distinct populations occupying specific anatomical domains.
Further analysis revealed the formation of cellular communities within the heart, where specific combinations of these cells are organized to perform the crucial functions of each heart region.
Together this dataset provides a high-resolution single-cell molecular and spatial cardiac cell atlas.
It also helps to explain how the distinct cell types of our hearts organize into the cardiac structures essential for the rhythmic ‘lub-dub’ that keeps us all alive!
Read the full issue of Omic.ly Premium 24

