The tumor microenvironment is a complicated place full of contradictions
Macrophages in lung cancer get their chance in the spatial omics spotlight.
A new single-cell study on non-small cell lung cancer is helping us understand why that is.
Lung cancer is the second most common cancer globally and the leading cause of cancer death.
Of the two types of lung cancer, non-small-cell lung cancer (NSCLC) is the most prevalent type, accounting for about 85% of all lung cancer cases.
Small-cell lung cancer makes up the other 15%.
Both major types of lung cancer are associated with smoking or exposure of the lungs to agents that cause DNA damage, although there are genetic risk factors that can make individuals more susceptible.
NSCLC includes several subtypes, but adenocarcinoma (LUAD) and squamous-cell carcinoma (LUSC) are the two that are seen most often.
But NSCLC isn't just a mass of cells growing uncontrollably, it's a complicated cellular environment that we're now just beginning to understand.
This tumor microenvironment (TME) has been the focus of cancer research for many years now and we’ve learned a lot about the roles of various immune cells (like T cells and B cells), in patient prognosis.
We’re also now appreciating the role of macrophages (eat bacteria and other bad things) in lung cancer progression.
Tumor-associated macrophages (TAMs) have been shown to perform paradoxical functions.
They can promote tumor growth by suppressing the immune response or they can inhibit it by promoting inflammation and engaging in cytotoxic activity against cancer cells.
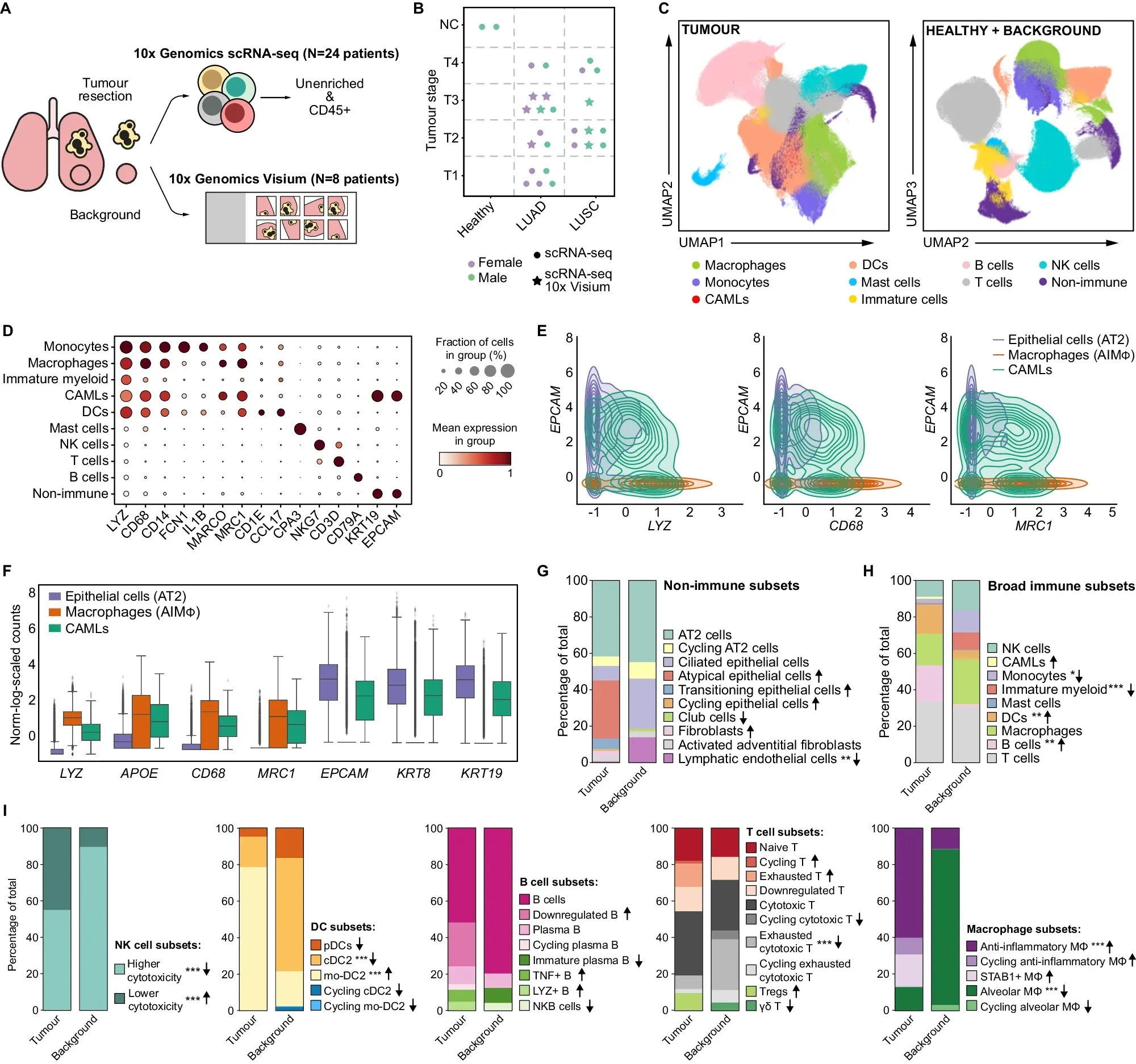
To better understand the role of these macrophages in NSCLC, the researchers behind today’s paper combined single-cell RNA sequencing (scRNA-seq) data from 25 patients with LUAD or LUSC with spatial transcriptomics from eight patients.
This combination allowed them to compare differences between tumor subtypes and also normal tissue.
What they found can be seen in the figure above: a) overview of the studies performed, b) umap comparing immune cell types found in normal and healthy tissue d-f) gene expression profiling of tumor associated immune cells g-i) further characterization of the state of the immune cells associated with healthy and normal tissues
This comprehensive TME profiling revealed important differences between the two subtypes of NSCLC.
The researchers found that each expresses different immune checkpoint proteins (inhibit the immune system’s ability to attack cancer) which means that they will likely respond differently to therapy.
This study provides a comprehensive dataset that identifies important molecular changes in the macrophage population within the tumor microenvironment, and could help guide therapeutic strategies against NSCLC.

