Sequencing proteins with nanopores is hard. Engineered nanopores might make it a little easier.
Nanopores, what can't they do? Well, they're not very good at sequencing proteins. Yet.
‘Why would anyone want to sequence a protein?’
Because, they’re the functional molecules in our cells and the ultimate end product of our genome!
And when our genome gets damaged, it’s changes in the amount of protein, when those proteins are made, how they’re modified or the creation of mutated proteins that results in disease.
So, knowing what proteins are present and how they’re broken is important.
Luckily, sequencing proteins can tell us something about all of those things!
There are a couple ways to approach doing this, but one of the newest and most interesting methods for sequencing things is to use nanopores.
Currently, nanopores are usually made using proteins that create tiny holes in a membrane.
As molecules pass through these holes, the size and the shape of the molecule changes the amount of electrical current that can pass through the hole.
This change in current can be measured, and those signals can then be used to predict what was passing through the pore at any given time!
Oxford Nanopore have been very successful in refining this technique to sequence both DNA and RNA.
But, proteins offer a slightly new challenge.
To get DNA and RNA to pass through a pore, a positive charge is placed on one side of a nanopore.
This draws nucleic acids through the pore because they have a net negative charge.
But proteins are different.
They are composed of both positively and negatively charged amino acids, so pulling them through a pore with a positive or negative electrical field would just end up with them getting stuck!
Fortunately, smart people realized that there are other ways to pull (or push) molecules through a pore, one of them being electroosmotic flow (EOF).
That might sound complicated, but it really just means: use water as the carrier.
And that’s exactly what the author’s of today’s paper did!
They had to get pretty creative though, because these pore proteins aren't good at EOF without a little help.
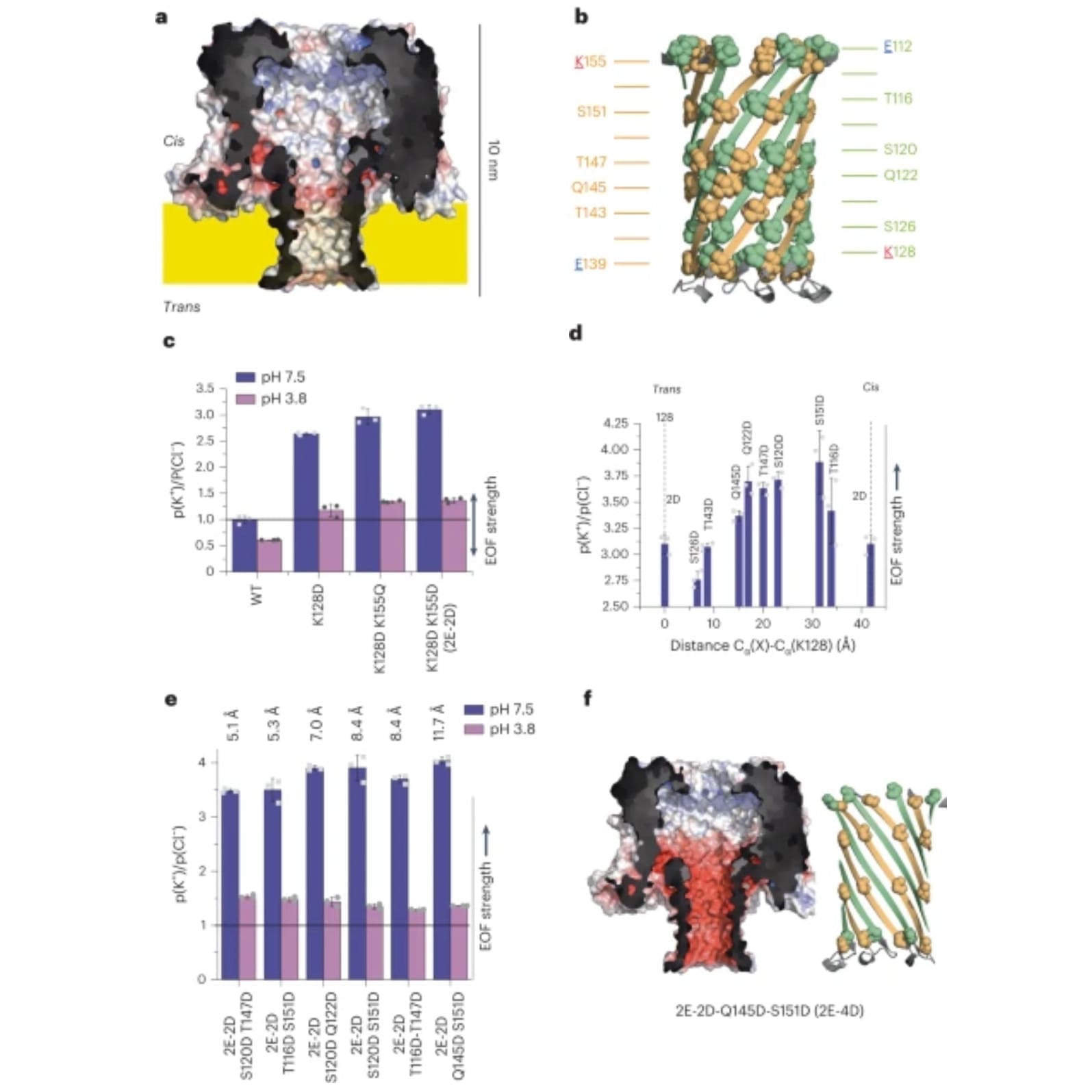
To do this, they specifically engineered a pore protein, CytK (a), and the barrel of the protein (b), to have negatively charged rings of amino acids to facilitate EOF when in the presence of an electric field.
The EOF performance of 2 (c), 3 (d), and 4 (e) rings was measured and it was found that 4 rings spaced 11.5 Å apart did the best.
The structure and charge of the winning pore, Q145D-S151D-CytK (2E-4D), can be seen in f.
Importantly, the authors went on to show that they can get proteins to pass completely through the pore (a fundamental requirement of any sequencer!) and generate current traces in the process.
But, converting that data into usable sequence is probably still many years out!
