Reverse Translation doesn't exist in nature, but that's not stopping us from trying to do it!
There are few things deserving of the superlative ‘holy grail,’ but Reverse Translation is one of them.
And that’s because it doesn’t exist in nature.
The Central Dogma states that DNA codes for RNA and RNA codes for protein.
But, proteins don’t serve as the template to code for other proteins, nor do they code nucleic acid.
We discovered in 1970 that some viruses can reverse translate RNA into DNA.
But the last bit about proteins has remained true since the Central Dogma was first postulated by Francis Crick in 1957.
That’s somewhat of a problem when it comes to being able to sequence proteins.
Because it means we don’t have any natural enzymes we can use to read back a protein sequence.
In DNA sequencing we have polymerases and reverse transcriptases that can spit out nucleic acid sequences, but we have nothing that can do that with proteins!
The current state of the art in protein sequencing uses ‘peptidases’ (enzymes that cut ‘peptide’ bonds) or Edman Degradation (a chemical reaction) that release amino acids at the end of proteins.
But in this scheme, detection is the hard part!
Some approaches fluorescently label cysteines and watch as the fluorescence signal decreases after every cycle of degradation.
This creates a cysteine sequence that can then be used to infer what protein was present.
Other approaches use antibody ‘recognizers’ to detect what amino acid is at the end of a protein before it’s degraded.
Neither of these approaches has scaled very well, they can’t detect every amino acid, and they struggle to recognize modified proteins.
With these issues in mind, the authors of today’s paper developed a method for performing reverse translation and sequencing of an immobilized peptide using DNA barcodes and antibodies.
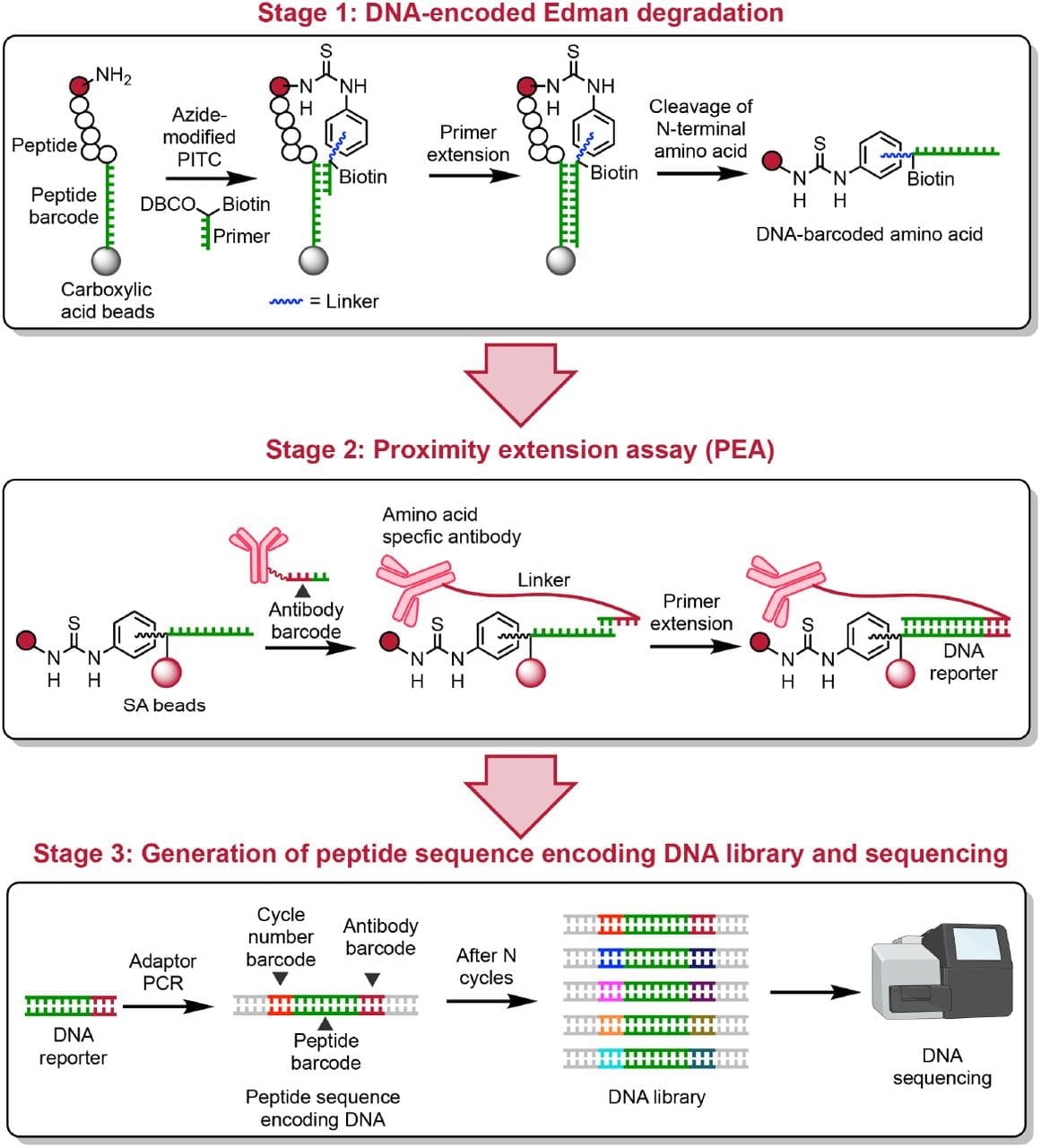
What they put together can be seen in the figure above.
Stage 1: A peptide is attached to a DNA barcode on its carboxy terminal end and exposed to a second oligo that binds the barcode sequence and covalently bonds to the amino terminal amino acid. A primer extension reaction is done to barcode the amino acid and then Edman Degradation is performed to release it from the complex. These barcoded amino acids are isolated every cycle.
Stage 2: The isolated barcoded amino acids are then exposed to oligo labeled antibodies that detect what amino acid is present. This adds an amino acid barcode and a cycle code to the DNA sequence.
Stage 3: The pieces of DNA from each ‘cycle’ of sequencing are combined, turned into sequencing libraries, and then sequenced on a DNA sequencer to determine the protein sequence.
The authors show that phosphorylation specific antibodies can be added at stage 2 to detect modified and unmodified amino acids.
While this process seems tricky to scale, it’s an interesting proof-of-concept that could be further developed into a real protein sequencer.

