Rare diseases get a second opinion and new diagnoses
A re-analysis of rare disease patients' genetic data delivers 500 new diagnoses
We've heard a lot recently about the utility of DNA sequencing in diagnosing patients with rare diseases.
And, thankfully, whole exome and whole genome sequencing are becoming the standard of care when a patient presents with a likely genetic disease.
The challenge here that still remains is that the diagnostic yield of sequencing a patient can vary greatly - from 20%-70%.
Some of this comes down to the techniques used, because not all sequencing is created equally!
Exomes target the coding regions of the genome (that aren't GC rich and don't have repeats), Short-Read whole genomes capture all of the variation that occurs in coding and non-coding regions but struggles to capture large genomic rearrangements (Inversions and translocations in particular, or mutations that happen in pseudogenes), and while long-reads provide the most comprehensive option for picking up genomic insults, they're still pretty pricey and just barely making their way into the clinic!
But the other issue at hand here when it comes to the variability we see in the solve rate is that when we sequence DNA, we find things, A LOT of things.
For example, you and I have, on average, 5 million single nucleotide polymorphisms (SNPs), 600,000 small insertions and deletions (indels), and 25,000 structural variants (SVs) in our genomes.
Of these, a few hundred will be "damaging" with unknown effects.
And the same is true for people who are afflicted with a likely genetic disease!
That is to say, finding a genetic cause for a disease really is like trying to find a needle in a haystack!
This also means that sometimes we miss disease causal mutations the first time we go digging through someone's genetic data and as informatics techniques improve, and we learn more about what mutations are associated with disease, we can go back and reanalyze old genomes to find new causes!
That's exactly what happened in this week's paper where researchers and clinicians from the Solve Rare Disease consortium (Solve-RD) re-reviewed genetic data from ~6,000 patients and 3,000 of their family members to come up with new diagnoses.
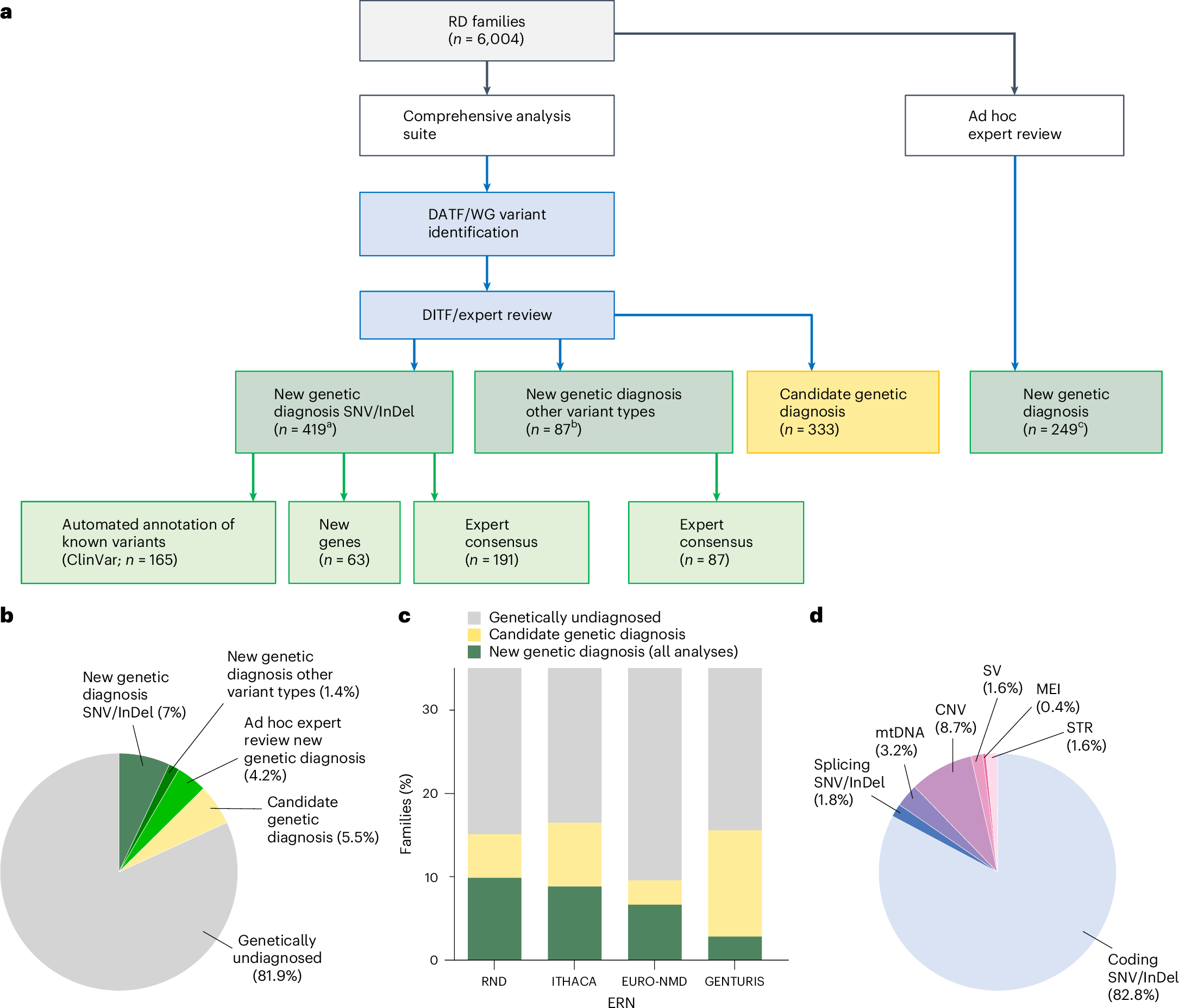
A summary of the results can be seen in the figure above where a) shows a breakdown of how the new genetic diagnoses were obtained b) a pie chart showing the diagnostic yield of this latest effort in the undiagnosed population c) bar chart showing yield per disease area: RND - rare neurological diseases, ITHACA - intellectual disability, telehealth and congenital anomalies, EURO-NMD - neuromuscular diseases, GENTURIS - genetic tumor risk syndromes d) Pie chart showing the breakdown of the disease causal variant classes
Overall, the re-analysis was able to provide a new diagnosis to 506 patients.
The hope here is that as we learn more, future analyses within this cohort will be able to turn up even more solves.
But since 95% of the patients in this analysis were exome sequenced and only 1.6% of the solves were from structural variants, maybe this cohort of undiagnosed patients would benefit from sequencing with more comprehensive sequencing methodologies that are better able to capture non-coding variation and structural variants.
