The story of the polymerase chain reaction and how sometimes timing is everything
The method below has been cited more than 600,000 times and is one of the most important developments in the history of science.
The polymerase chain reaction (PCR) is used widely to amplify DNA sequences.
Kary Mullis is often given sole credit for developing PCR, which, according to him, he discovered while "riding DNA molecules" during an acid trip in the early 1980's.
But, today's story begins a decade earlier in the lab of Har Gobind Khorana.
Khorana won the Nobel Prize in 1968 for his work figuring out how RNA codes for protein.
The key to his success was that he and his team synthesized their own molecules of RNA, called oligonucleotides.
This allowed them to see what amino acids ended up in proteins after the translation of their sequences.
Khorana continued to focus on scaling this synthesis process to create a full gene sequence.
One of his post-docs, Kjell Kleppe, had an idea to use little pieces of complementary DNA to kickstart these synthesis reactions and copy a target sequence with DNA polymerases.
At the time, it was known that polymerases were involved in copying DNA during cell division and could use a 'primer' to start this reaction.
He presented his method and initial data in 1969 and published a paper where his method of in vitro "DNA repair replication" was described in 1971.
So, why does Mullis get all of the credit for the discovery of PCR?
Mostly because of timing.
In 1970, the Khorana lab was one of the few groups making oligonucleotides.
Kleppe's result was seen as interesting but technically infeasible to scale and the true power of DNA copying wasn't recognized until the early 1980's when cloning and other molecular manipulations were really taking off.
Mullis was employed at that time by Cetus Corporation and he stumbled on the idea of copying DNA using primers and polymerases all on his own.
He published an initial paper on PCR in 1985 using the E. coli ‘Klenow’ fragment (a truncated portion of DNA polymerase) which required manually cycling the reaction in a water bath and adding back enzyme every round because it wasn’t heat stable.
However, Mullis realized he could use the polymerase from a different bacteria, T. aquaticus, which lives in the boiling hot springs of Yellowstone Park.
This polymerase, which we now call Taq, was heat stable and didn't require replacement between cycles!
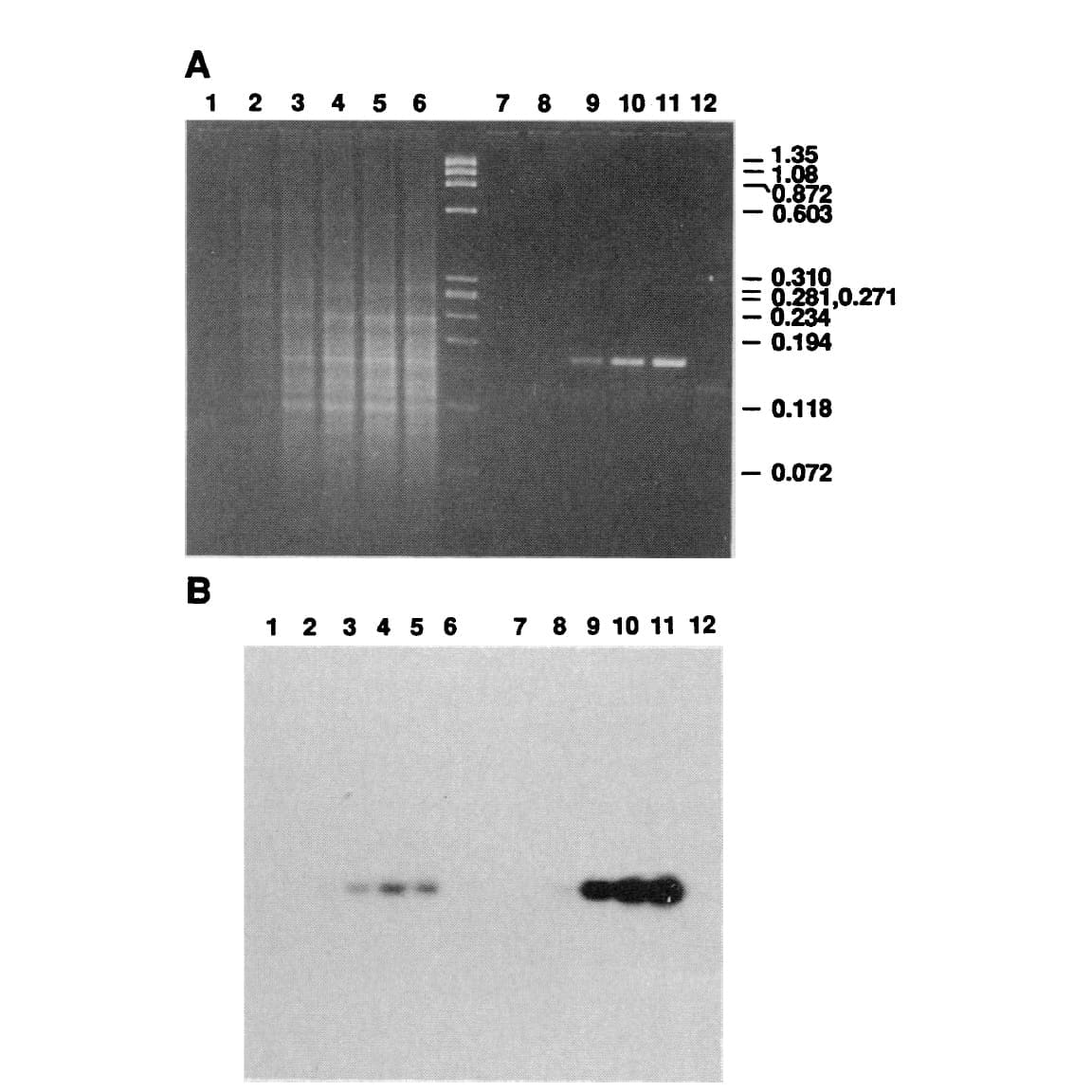
The figure above shows Mullis' comparison of Klenow (Lanes 2-5) to Taq (Lanes 8-11).
(A) is an agarose gel and (B) is a southern blot using a radioactive probe to detect the targeted DNA.
The benefits of using Taq are pretty obvious, but its heat stability was game changing because it made the process much cleaner and easy to automate!
Mullis received the Nobel Prize for this work in 1993.
