Omicly Weekly 9
January 28th, 2024
Hey There!
Thanks for spending part of your Sunday with Omicly!
This week's headlines include:
1) The evidence that Epstein-Barr Virus can cause multiple sclerosis is mounting
2) The importance of taking out the trash: An overview of cellular protein degradation pathways.
3) Linus Pauling proposed a triple helical structure for DNA in 1953. Here's why he got it so wrong.
Please enjoy!
It has been hypothesized that infection with Epstein-Barr Virus can result in the development of multiple sclerosis. We now have good evidence that this is true.
But you might be thinking to yourself, “Epstein-Barr? Isn’t that the virus that causes mono? WE’VE ALL BEEN INFECTED WITH THAT!”
While it’s true that 95% of people have been infected with Epstein-Barr (EBV), only about 1 in 300 people in the United States have multiple sclerosis (MS).
Recent research has shown that genetic risk for MS emerged in northern europe 5,000 years ago, providing some evidence for why MS affects predominantly people of northern european descent.
The hypothesis being that as humans switched from a nomadic to a pastoral lifestyle, MS associated variants, mostly in the HLA genes, conferred an evolutionary advantage to these people.
And because MS is a disease of later onset, ages 20-50, these variants did not negatively impact their ability to reproduce.
But the impact here on the HLA genes is important, because HLA helps our immune system detect and clear infections!
HLA is also implicated in autoimmune diseases like Lupus, Psoriatic Arthritis and MS where the immune system inappropriately identifies its own proteins as foreign.
In the case of MS, it attacks the proteins that make up the protective coating (myelin) of neuronal cells.
This de-mylelination is a hallmark of MS and results in many of the symptoms associated with the disease including muscle weakness, eye pain, and loss of coordination.
But, you’re probably asking yourself what this all has to do with EBV?
Well, large population studies in the early 2000’s identified that EBV infection appeared to be a risk factor for developing MS.
And more recent studies have shown that the antibodies that recognize the EBNA-1 protein from EBV also cross-react and recognize the ‘glue’ that holds neuronal cells together!
But this presents an interesting question: If antibodies to EBV also recognize a neuronal protein, why doesn’t everyone that’s infected with EBV develop MS?
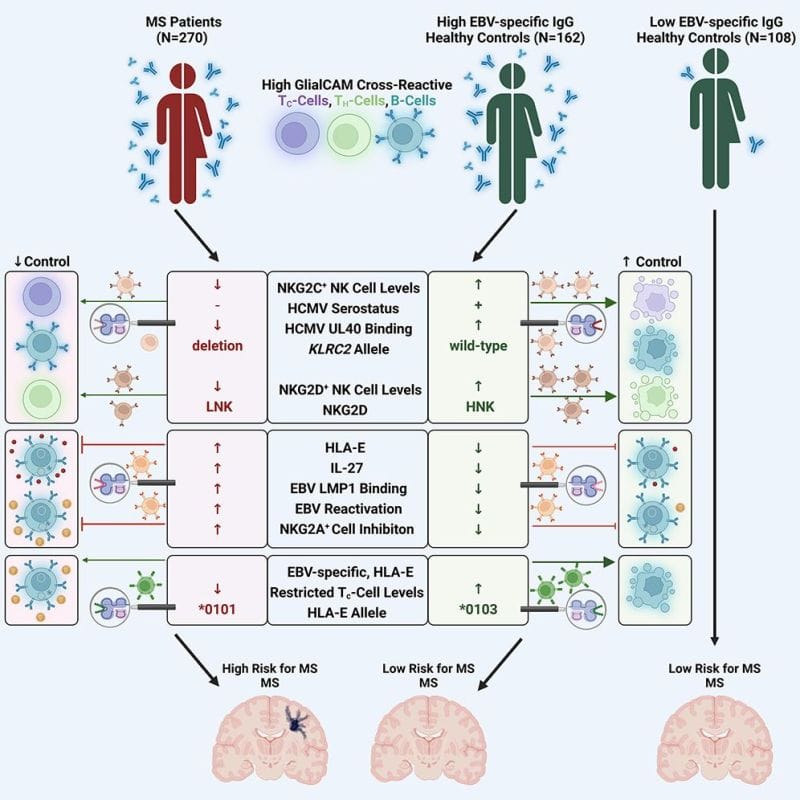
To answer that question, the author’s of today’s paper looked at differences in the immune systems of people infected with EBV who did or did not display symptoms of MS.
What they found was super interesting.
It turns out that a number of factors are at play here and that the controls that are in place to prevent autoimmunity fail.
This failure also seems to be closely associated with HLA-E expression in immune cells.
And, people with elevated levels of HLA-E had up to a 260 fold increased risk of developing MS!
While EBV may not be the only contributing factor in the development of MS, its association with MS could provide new pathways for the treatment or prevention of the disease.
###
Vietzen H et al. 2023. Ineffective control of Epstein-Barr-virus-induced autoimmunity increases the risk for multiple sclerosis. Cell. DOI: 10.1016/j.cell.2023.11.015
The importance of taking out the trash: An overview of cellular protein degradation pathways.

This might not be obvious, but cells generate a lot of garbage.
Like anything, parts of cells get damaged or worn out and eventually need to be replaced.
This is true of small proteins all of the way up to mitochondria or other large organelles.
Some cells, like those in our immune system, even EAT THINGS like bacteria or viruses which are really just juicy bags of protein that need to be degraded!
But it’s not only broken proteins or microbes that get disposed of.
During development, cell division, or even throughout the day, a cell’s needs change.
This means that the proteins that are present also need to change!
This is especially true when cells ‘differentiate’ into all of the different cells that make up our organs and tissues.
And, if you remember, YOU are the product of one sperm and one egg that divided into trillions of different cells!
To get from a fertilized egg to YOU, changes in protein content within all of those cells needed to be made.
And the changing protein needs of cells are, in part, regulated by protein degradation pathways!
There are two major ways to chop up proteins in a cell:
The Proteasome - These are like protein wood chippers. They’re used to help with the rapid proteolysis (big word for chopping) of useless, misfolded, damaged, or no longer needed proteins!
The proteasome is itself a protein complex that recognizes a specific tag on proteins marked for destruction and dices them up into ~7 amino acid chunks.
Those chunks can then be further degraded and reused to create new proteins!
The tag that the proteasome recognizes is a 76-amino acid protein called ubiquitin.
And this tag is added by a grim reaper-like protein called ubiquitin ligase.
Once a chain of at least 4 ubiquitins is added to a protein, it is recognized by the proteasome and gobbled up.
The Lysosome - If the proteasome is a wood chipper, then the lysosome is like an industrialized garbage dump.
The lysosome is a specialized membrane enclosed organelle (kind of like a bathroom inside a house) where bulk protein degradation activities take place.
They’re filled with proteases (small protein chomping enzymes) and have a highly acidic environment to aid in the breakdown of whatever is unfortunate enough to end up in there.
The lysosome is used for digesting nutrients that have been brought in from outside the cell, degrading proteins that have hung around for too long, disassembling and recycling major cellular organelles like mitochondria (autophagy), or bulk destroying proteins during times of stress.
And it's probably not surprising that problems with both the proteasome and the lysosome are implicated in many diseases.
Because, proteostasis, or the maintenance of the proteome through the creation and destruction of proteins, is key for normal cellular function!
The kaleidoscopic image below is the triple helix Linus Pauling proposed as the structure of DNA in February 1953. Here's why he got it so wrong:
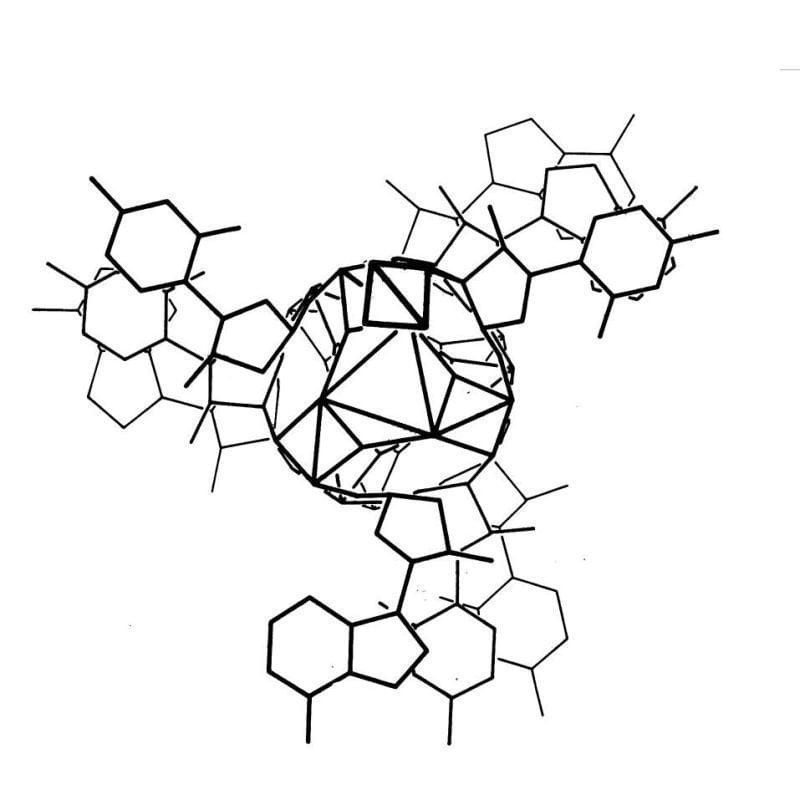
Linus Pauling is remembered as one of the greatest American scientists of all time. He was awarded two undivided Nobel Prizes, and is basically the father of modern structural biology.
He's most famous for solving the 3D structure of the α-helix and the β-sheet, the two most common protein structures.
To do this, Pauling employed a technique called x-ray crystallography which basically bombards a crystal with x-rays. How those x-rays bounce off of a crystal tells you something about the shape of the item that was crystallized and the resulting 'diffraction patterns' can be used to determine key structural features of the crystallized molecules.
In his work, Pauling took this a step further and was one of the first scientists to use that information to model molecular structures with balls and sticks.
Fresh off of his foundational work with proteins, Pauling turned his attention to DNA. While Pauling famously thought that DNA was NOT the genetic material, there's a rumor that he heard that teams in the UK were close to solving DNA's structure and the ever competitive Pauling wanted to see if he could beat them.
However, Pauling didn't generate his own data and relied heavily on data and calculations derived by others in the field, namely those created by William Astbury and Florence Bell in 1938.
Unfortunately, Pauling was unaware of the most recent advancements in the diffraction of DNA by Franklin and Gosling.
But he did have access to electron micrographs of DNA and the 15 year old Astbury/Bell data which indicated the structure was helical.
Based on incorrect density calculations, he also proposed that the structure had 3 nucleotides at each position.
It was already known that DNA was composed of a sugar phosphate backbone, so all of these things taken together led Pauling to a triple helix.
To generate the image above, Pauling and Robert Corey placed the phosphate backbones of 3 DNA strands at the center of the structure with the nucleotides facing out, like the spokes of a bicycle wheel.
In hindsight, we know this is totally wrong and there are 3 key errors that make this structure implausible:
1. At pH 7, the phosphate backbone is negatively charged so 3 of them packed together would repel one another
2. The model doesn't leave space for sodium and Astbury's diffractions were all sodium salts
3. Chargaff's nucleotide pairing rules were totally ignored
Fortunately, taking inspiration from Pauling and his use of modeling, Watson and Crick, armed with pristine x-ray diffraction data from Franklin and Gosling, published the correct double helical structure for DNA two months later in the April 1953 issue of Nature.
###
Pauling L, Corey RB. 1953. A Proposed Structure For The Nucleic Acids. PNAS. DOI: 10.1073/pnas.39.2.84
