Omicly Weekly 6
January 7, 2024
Hey There!
Thanks for spending part of your Sunday with Omicly!
This week's headlines include:
1) Pfizer developed a technique for determining the primary structure of their mRNA vaccine including detecting base modifications, the 5'-cap, and the poly-A tail!
2) The Proteome: what it is and why protein structure is so important
3) The story of Phoebus Levene, one of the mostly invisible grandfathers of nucleic acids
Please enjoy!
Pfizer developed a new method to determine the primary structure of RNA. 'So, they made a sequencer?' Kinda.
Two of the most widely used vaccines during the pandemic were based on mRNA technology.
Messenger ribonucleic acid (mRNA) is the template that's used by our cellular machinery to create proteins.
Traditionally, vaccines are made using killed viruses, weakened viruses, or viral proteins that have been produced synthetically.
These all make great vaccines, but creating vaccines this way is time consuming, and that's something that's in short supply during a pandemic!
This is where mRNA comes in and this technology leverages decades of research on how best to express proteins directly in cells, and in the case of a vaccine, present those proteins for recognition by the immune system.
Getting mRNA into cells is relatively easy.
But that's when the problems start because our cells are very sensitive to RNA and they chop up anything they think is foreign as a defense mechanism.
Thankfully, it was discovered that if you replace uracil in the mRNA with a modified base, N1-methyl-pseudouridine, cells didn't sound the invasion alarm.
But that's not the whole story because for an mRNA to be stable and translated into protein, it must be 5'-methylated or capped, and also have a Poly Adenosine (Poly-A) tail.
So, for an mRNA vaccine to work, it needs to be capped, have a poly-A tail and be full of modified uracils.
How are we sure that the mRNA in the vaccine has all of these things?
I imagine that in the fall of 2020 a series of emails were exchanged with the FDA.
FDA: What quality control have you done on the primary structure of the RNA to be sure it's translation competent?
Pfizer: We sequenced it, it's fine.
FDA: Thank you for your reply. Is the cap intact? What about the poly-A tail? And you used N1-methyl-pseudouridine instead of uridine (uracil), how do you know it's N1-methyl-pseudouridine in there?
Pfizer: ...
At least this is how I'm guessing it all went down.
So Pfizer had to figure out a way to show that everything about the primary structure was correct. Not just the sequence but that the sequence also contained all of the appropriate modifications.
They decided to do this with mass spectrometry because it can be used to determine the chemical makeup of molecules in a solution.
It does this by ionizing the molecules and then smashing them into a detector to determine their mass to charge ratio (pretty cool!)
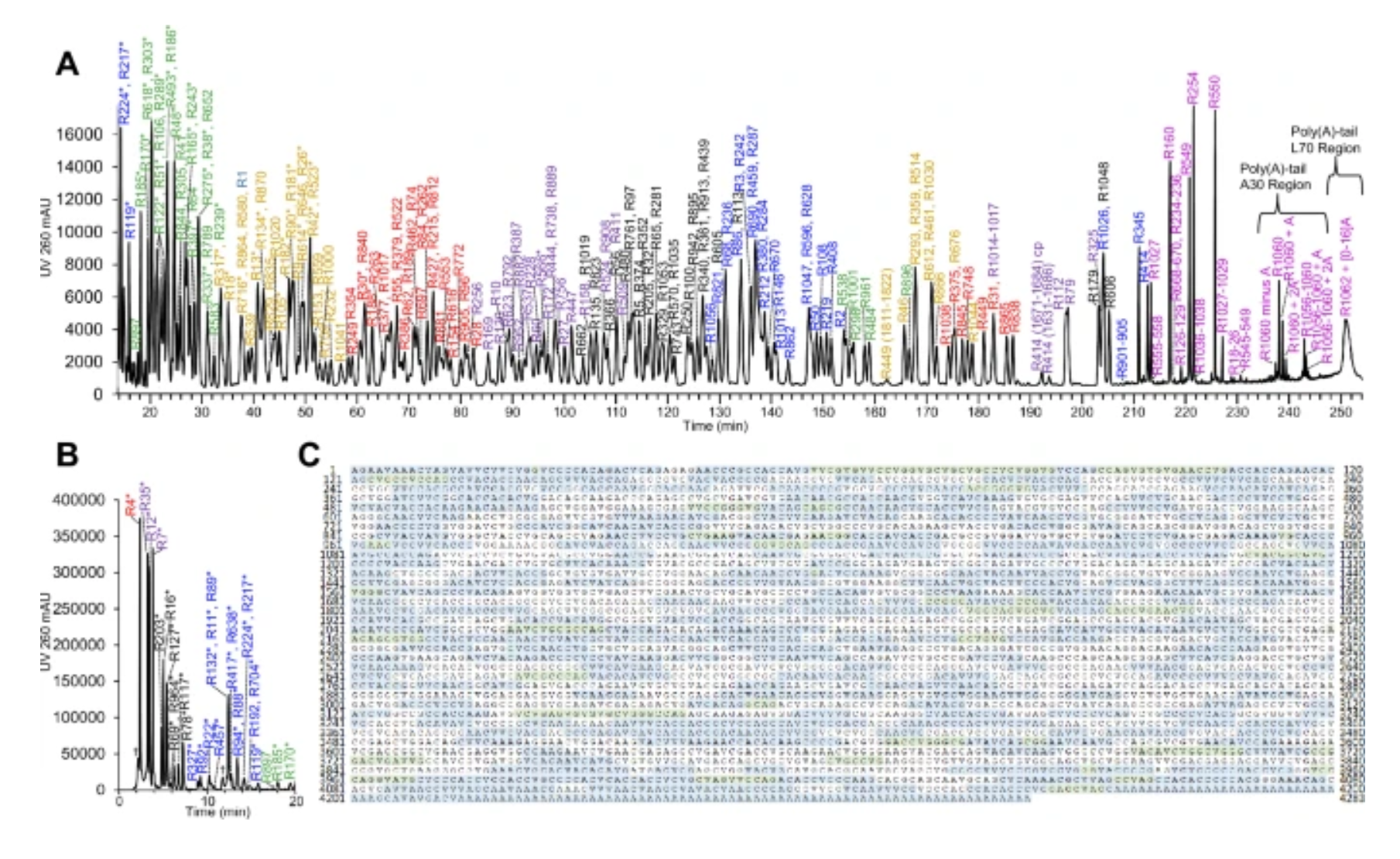
And so that brings us to the figure below.
Here Pfizer chopped up their vaccine mRNA, threw it on a mass spec, and looked at the masses of everything that came out making sure they detected all of the sequence features including the methylated cap and the poly-A tail!
This is an ingenious way to determine the complete primary structure of a complex nucleic acid beyond just sequencing its bases!
###
Gau BC et al. 2023. Oligonucleotide mapping via mass spectrometry to enable comprehensive primary structure characterization of an mRNA vaccine against SARS-CoV-2. DOI: 10.1038/s41598-023-36193-2
The Proteome: we can’t live without it, but, what actually is it?

It’s pretty simple:
The proteome is all of the proteins that are coded for by your DNA!
However, the process of creating proteins is somewhat complicated and to get to those beautiful, functional, molecules, we actually need to start by talking about the ‘central dogma.’
The central dogma of molecular biology was first theorized by Francis Crick in 1957.
It states that DNA codes for RNA which codes for proteins and once a protein is made, there’s no going back!
You can’t code for RNA, DNA, or other proteins from protein!
Proteins are made through the ‘translation’ of messenger RNA.
This is a process that involves a large protein complex called the ribosome which ‘reads’ the RNA and stitches together protein building blocks (amino acids) to create the final molecule.
But why are proteins so important?!?
Because they’re the molecules in our cells that do all of the work!
They:
1) Form the structural components of our cells
2) As enzymes, they perform the complex chemistry that makes life possible
3) They also do cool things like copy DNA, read RNA to create other proteins or serve as signaling molecules!
Since proteins perform basically all of our important cellular functions, their presence, absence or altered performance is what defines disease.
Presence and absence are pretty straightforward:
If a protein is needed and it’s not there…that’s bad.
If a protein isn’t needed and it's there…that’s also bad.
But altered performance might be something that’s a little tougher to wrap your head around, so let’s talk about how proteins actually work!
Something we mention A LOT in molecular biology is that structure equals function.
The structure of most proteins is highly complex and we define protein structure across 4 levels:
Primary Structure - Sequence of the amino acids that make up the protein.
Secondary Structure - 2-dimensional structure of the protein. This includes the local interactions of amino acids with one another through hydrogen bonds and disulphide bridges. The most common secondary structures are the α-helix and β-sheet.
Tertiary Structure - Overall 3-dimensional structure of a protein that forms once all of the secondary structures fold into their final forms and interact with one another.
Quaternary Structure - Formed through the interactions among multiple proteins to create a functional protein complex. Many proteins work together with friends to do their jobs!
‘Ok, cool story, but what does any of that have to do with the proteome?!’
Because proteomics is the study of how all of the proteins in your cells and tissues interact to perform their cellular functions!
And understanding how those things break, including the impact on protein structure, function or interactions with their friends, is how we can use proteomics to predict and prevent disease!
Watson and Crick were the first to describe the structure of the DNA double helix. Their major contribution to science was, at its core, a synthesis of all of the best data available at the time.
This statement isn’t meant to discount what they did in 1953, but more so to recognize all of the foundational work that preceded them.
1869 - Nuclein (DNA) is discovered - F. Miescher
1912 - The hydrogen bond is proposed - T. S. Moore and T. F. Winmill, Popularized by L. Pauling
1935 - DNA is a macromolecule with a sugar phosphate backbone - P. Levene
1943, 1952 - DNA is the genetic material - Avery-MacLeod-McCarthy, Hershey-Chase
1950 - A and T, G and C are in a 1:1 ratio - E. Chargaff
1952 - DNA is a helix - Rosalind Franklin, she got the math right
1953 - Watson-Crick base pairing - Broomhead, Donahue, Watson, Crick
While all of these pieces were required for the final structure to be determined, the bulk of the basic chemistry was done between 1905 and 1935.
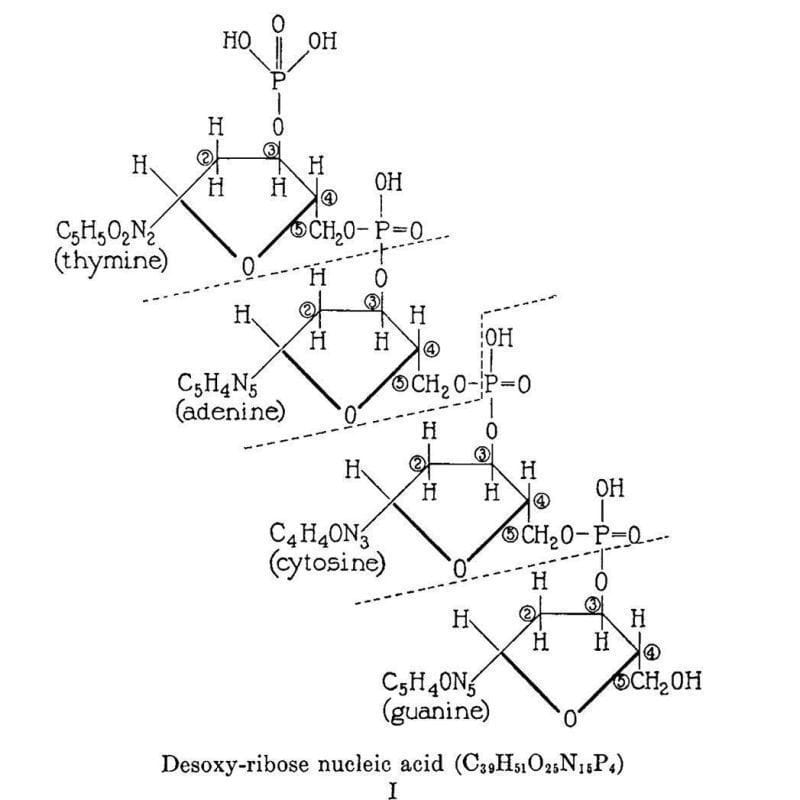
The figure above is the culmination of the life’s work of Phoebus Levene.
He's probably not someone you’ve heard of before.
He was the first to discover that nucleic acids are composed of ribose sugars.
He also determined that these sugars were connected by a phosphate backbone.
And, he eventually figured out that DNA is made of 2-deoxy-D-ribose while RNA is D-ribose.
Oh, he also confirmed the identities of all of the bases: Adenine, Thymine, Guanine, Cytosine and Uracil.
And he's the reason why we call these bases nucleotides!
Pretty mind blowing, right?
This work is summarized in the figure below which displays Levene's 2 dimensional chemical structure for 'Desoxy-ribose nucleic acid.' This structure isn't perfect (Cytosine is labeled as a diphosphate and the chemical composition of the nucleobases is a little off) but it does accurately depict the 5'-3' sugar phosphate backbone of DNA.
Now, the reason why you've never heard of Levene is because he proposed the 'tetranucleotide hypothesis' which stated that DNA was merely a structural molecule composed of 4 repeating nucleotides.
He was certain this meant DNA was too simple to be the carrier of genetic information.
Like everyone else, he believed that proteins were the obvious choice as the genetic material since there are 5 times as many amino acids.
Unfortunately, Levene's hypothesis took the spotlight off of DNA for decades and this has been described as a 'scientific catastrophe' despite Levene getting the basic chemistry and the 2D structure of DNA mostly correct.
So, for Watson and Crick to finally put the 3D puzzle together it took a renewed interest in DNA as the genetic material, Levene's chemistry, the ratio of the bases from Chargaff, (borrowed?) X-ray diffraction data from Franklin, and a couple pints at the Eagle pub in Cambridge.
The rest is (a very disputed) history.
###
Levene PA, Tipson RS. 1935. The ring structure of thymidine. J. Biol. Chem. DOI:10.1016/S0021-9258(18)75193-4
