Omic.ly Weekly 12
February 18th, 2024
Hey There!
Thanks for spending part of your Sunday with Omic.ly!
This week's headlines include:
1) The yeast used to brew Guinness is unlike any other found in Ireland or the UK
2) Protein sequencing is the latest cutting edge technique in our proteomics toolkit
3) Sickle Cell Anemia was molecularly characterized in 1949 using a revolutionary new technique: electrophoresis
Please enjoy!
The yeast used to make your most recent Guinness isn’t like any other found in Ireland or the UK!
Not surprisingly, beer brewing seems to have gotten its start at the same time as the domestication of rice and cereals.
The fermentation of those grains came soon after and were mostly a family affair with small personalized brews being made in clay pots with the aid of airborne yeast.
The majority of these ancient ‘breweries’ have been found in the middle-east and China dating back as far as 7,000 BCE!
But the modern version of beer that we think of today didn’t arrive on the scene until the 14th and 15th centuries when the process became more common with the addition of hops (Yay Germany!), saving cultures, and the establishment of Pubs and Monasteries that routinely served the beverage.
Beer making really became industrialized in the late 18th century with the invention of the thermometer and hydrometer which allowed for better control of the brewing process.
This also made the process much more scientific!
And science is (mostly) responsible for all of the different types of beer that we have today.
Many of these arose in specific geographic regions with different flavor profiles originating from the malt and hops endemic to the region doing the brewing.
But there’s one other major contributor to the flavor of beer:
Yeast!
While the raw materials that go into the beer making process are important, it’s the yeast that eats those materials to create the ethanol and the metabolites that form much of the flavor profile of our favorite brews.
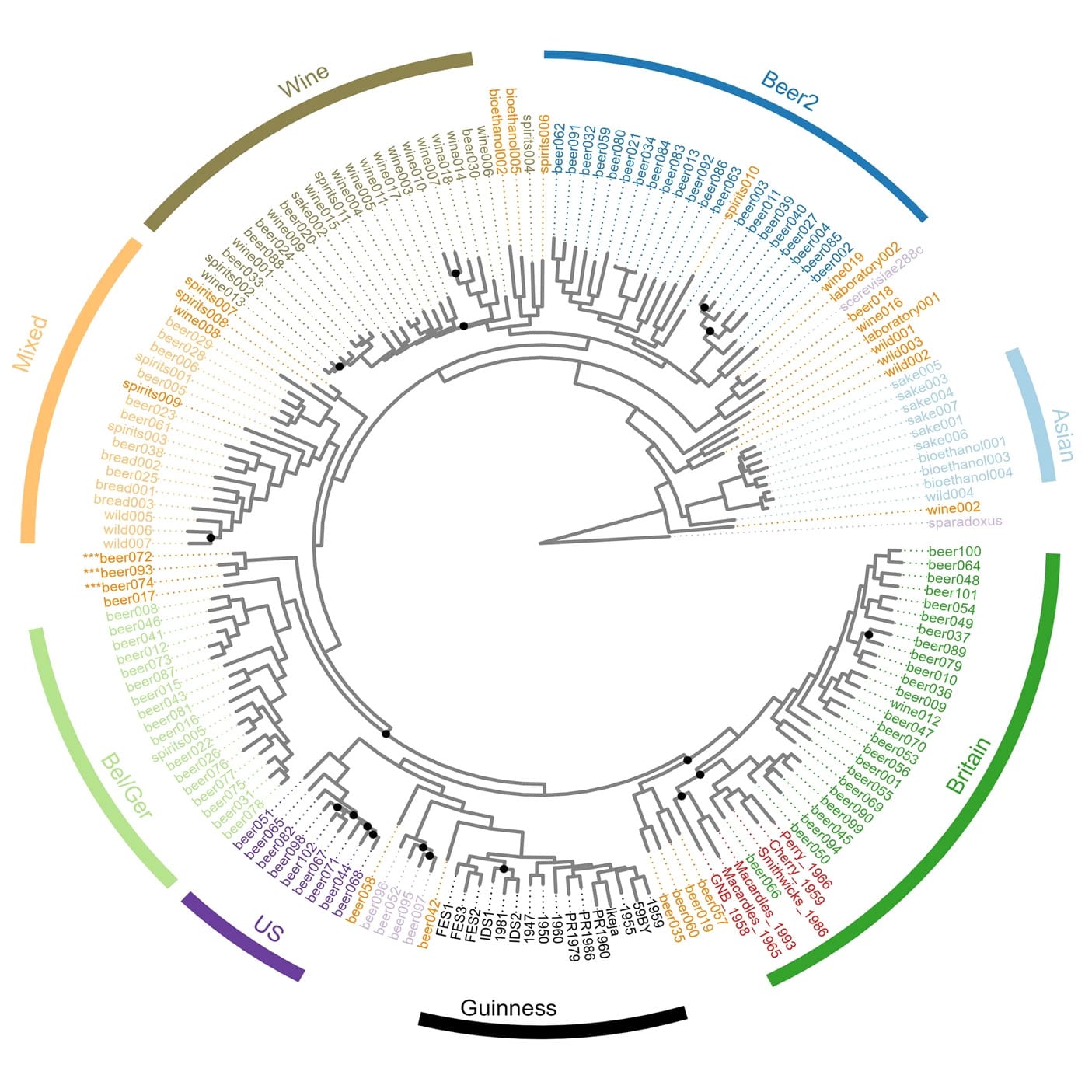
Guinness is no exception and the researchers behind today’s paper sought to identify the origins of the yeast used to brew this iconic and velvety stout.
The Guinness brewery was established in 1759 and maintains detailed records of its brewing process.
It knows that the yeast it uses today is one that originated from a 1903 Watling Laboratory Guinness yeast.
However, the origin of THAT yeast is unknown, so the researchers used both short-read and long-read whole genome sequencing to both confirm the identity of Guinness yeast and find its next closest ancestor!
What they found was surprising and their analyses revealed that Guinness yeast isn’t related to any Irish or British strains.
Its closest cousins are actually US yeasts!
And its closest ancestor is Beer042, an isolate that was likely obtained from the Carlsberg group yeast collection, and used to brew a Belgian lager style beer.
But, the Guinness yeast strain is special and shows a mosaic ancestry, sharing only about 40% genetic identity with Beer042.
The authors believe this is due to how the Guinness yeast was handled and used in the early days which resulted in distinct genetic adaptations that gave rise to the beer (and yeast) we all know and love today!
###
Kerruish DWM et al. 2024. The origins of the Guinness stout yeast. Nature Comm. DOI: 10.1038/s42003-023-05587-3
Protein sequencing is the latest cutting edge technique in our proteomics toolkit. But when will it be ready for primetime?

The central dogma states that genetic information flows from DNA -> RNA -> Protein and that proteins don’t code for anything.
Organisms have evolved methods to make DNA from DNA and DNA from RNA (viruses!), but there are no enzymes that work backwards from proteins!
This makes the job of figuring out how to sequence proteins very hard because there aren’t any naturally occurring enzymes that we can co-opt to do our bidding like we can for DNA and RNA sequencing.
Luckily, we’re surrounded by creative scientists who are trying to defy nature and tackle this problem head on:
Fingerprinting (Nautilus Biotechnology) - Use a flowcell to bind and immobilize billions of full length proteins and expose them to different types of affinity reagents to sequentially probe their structures. The detection of 'features' at each position can be used to infer what protein is present and affinity reagents that can broadly recognize post-translational modifications (PTMs) are in development.
Edman Degradation (Erisyon) - A method where peptides are immobilized on a glass slide, cysteines are fluorescein labeled and then the slides are subjected to multiple cycles of chemical degradation. The fluorescence at each cycle is recorded and the position of the cysteines within each peptide can be inferred by the reduction of fluorescence every cycle. The sequence of the cysteines can then be used to infer which peptide was present.
Kinetic Sequencing (Quantum-Si) - Proteins are sequenced using fluorescent 'recognizers' that bind to the ends of peptides. Recombinant proteases are used to sequentially expose the next peptide in the chain for recognition and sequencing. The binding kinetics of the 'recognizers' can be used to both predict the terminal amino acid and whether it is modified with a PTM.
Nanopore Sequencing (ONT) - Oxford Nanopore and others are working on modifying nanopores to directly sequence proteins. This is similar to RNA and DNA applications where the molecules are linearized and pulled (or in the case of proteins, pushed) through the nanopore’s channel for detection. We’re still in the very early days on this one so it might be a minute before we see these in the hands of researchers!
Reverse Translation (Encodia) - Encodia are mostly a stealth mode company and have released very little information about how their technology works. But, they refer to it as reverse translation, (again, this doesn’t exist in nature), and their website describes using oligo-tagged ‘affinity reagents’ (Antibodies? Aptamers?) to sequence proteins.
Despite the fact that protein sequencing is still in its infancy, I’m excited to see what these and other companies are able to come up with to help reveal the secrets hidden in our proteomes!
Sickle Cell Anemia was the first inherited disease to be molecularly characterized. It was done in 1949 using a revolutionary new method: electrophoresis.
Sickle cell anemia affects 4.4m people, and 43m are carriers of the trait.
It is characterized by the crescent, or sickle shape, of the red blood cells of those affected by the disease.
James Herrick first discovered sickle-shaped blood cells in a patient suffering from severe anemia in 1910.
Through subsequent observation it was realized that there was an asymptomatic form of the disease, sickle cell trait.
In those individuals it appeared that they had a mixture of normal and sickle blood cells.
Further study within the families of these individuals in 1923 revealed that sickle cell was hereditary or passed down from parents to their offspring.
And because those with sickle cell trait appeared to have a 50/50 mix of sickle/normal blood cells, it was determined that this was a recessive Mendelian disease.
Linus Pauling, a titan of early molecular biology, was no stranger to blood or the protein hemoglobin and spent many years in the 1930’s studying hemoglobin’s interactions with oxygen.
Pauling had a suspicion that the structure of proteins played a vital role in their function and was first introduced to sickle cell anemia in 1945.
He hypothesized that the sickling of cells could be related to a change in the structure of hemoglobin since red blood cells are literally just bags that contain a boat load of hemoglobin protein.
So he and his team, Harvey Itano and John Singer, tried to figure out a way that they could show that a difference in the structure of hemoglobin was the cause of sickle cell anemia.
After a bit of trial and error, they stumbled on the use of electrophoresis, a brand-new technique at the time, that allowed for the separation of molecules based on their electrical charge.
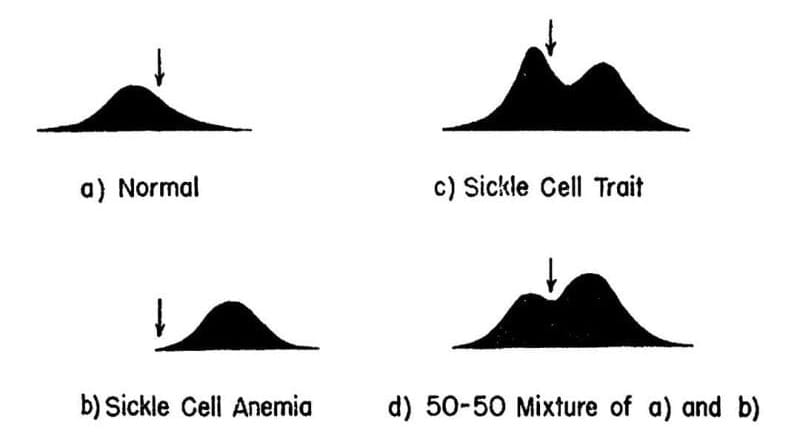
The results of their experiments can be seen above. The team separated and quantified 4 sets of blood samples using Longsworth scanning diagrams. A) shows normal hemoglobin, B) is hemoglobin from a sickle cell patient, C) is hemoglobin from a patient with sickle cell trait, and D) is a mixture of A and B. The arrow denotes a point of reference for comparing the diagrams.
This work demonstrates that there is a molecular basis for sickle cell anemia and that changes to a gene can alter the structure of a protein.
In the case of sickle cell, this functional relationship extends further because in the 1950’s, the trait was shown to be protective of malaria.
This explains evolutionarily why this disease is found in individuals of African descent; however, it fueled an unfounded fear of 'black blood' throughout the early 1900's.
###
Pauling L et al. 1949. Sickle Cell Anemia, a Molecular Disease. Science. DOI: 10.1126/science.110.2865.543
