Omicly Weekly 11
February 11th, 2024
Hey There!
Thanks for spending part of your Sunday with Omicly!
This week's headlines include:
1) 80% of patients with an autoimmune disease are women, we are now beginning to understand why
2) Mass Spec is leading the charge in bringing high-throughput proteomics to a lab near you
3) The "most beautiful experiment in biology" was performed by Meselson and Stahl in 1957
Please enjoy!
Approximately 80% of people with autoimmune diseases are women. We may now understand why this extreme gender bias exists.
You're probably aware that biologically female individuals have two X chromosomes.
You might not be aware that one of those X’s is turned off in every cell in a female’s body.
The reason for this is that the X chromosome is full of genes!
It’s so important that every human needs at least one to live, and that includes the boys!
But, too much of a good thing can be a problem, and in females, one of these X’s is turned off in a process known as X-inactivation.
The key molecular mechanisms behind this process were finally revealed in the early 90’s.
And X-inactivation is the work of a long non-coding RNA, the X-inactive specific transcript, or Xist for short.
Xist is able to turn off one of the X’s by recruiting a bunch of RNA binding proteins to itself which in turn causes the X that expresses Xist to shrivel up into a little ball that can’t be accessed for gene expression.
It’s a pretty elegant solution to the two X problem!
But you might be asking what this has to do with autoimmune diseases?
Well, they’re predominantly a female problem, and it’s especially bad in Lupus where 90% of patients are female
The conventional thinking was that since autoimmune diseases usually appear when women reach child bearing age, they were the result of hormonal changes.
But, the researchers in today’s paper hypothesized that X-inactivation also contributes, and that antibodies against the Xist RNA binding proteins play a role in the development of disease.
This isn’t a crazy idea!
It has been known for some time that many patients with autoimmune diseases have high titers of antibodies against RNA binding proteins!
To test this hypothesis, the authors used a mouse strain where females, and not males, are very susceptible to developing a lupus-like disease after exposure to a chemical (Pristane).
But, to test whether this susceptibility is the result of autoimmunity to RNA binding proteins and X-inactivation, they engineered MALE mice to carry a fake chromosome where they could control the expression of Xist with a drug (doxycyclin)!
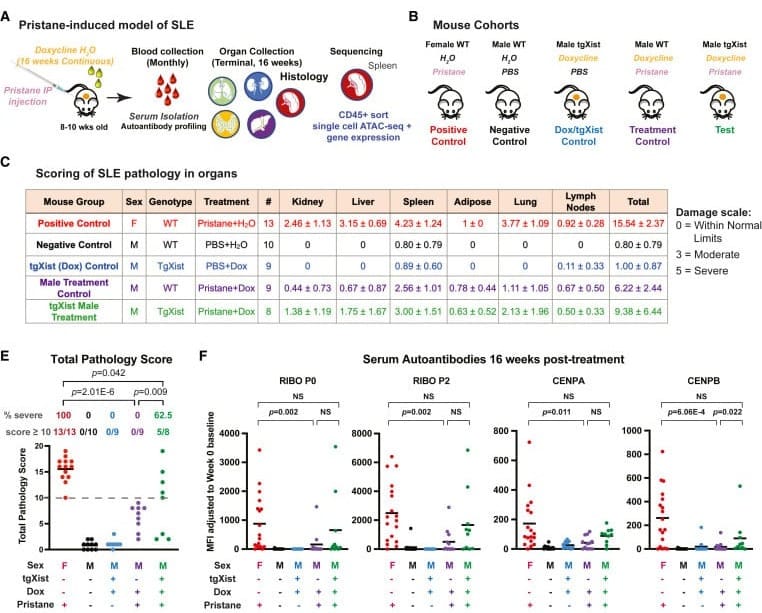
The results of this experiment can be seen in the figure below. A) Is an experimental overview, B) Explains the different treatments, C) Shows similar levels of organ damage in treated females and males where Xist was induced, but not in controls, E) Is a graphical summary of the table, and F) Measures the presence of Lupus associated autoantibodies.
Taken together, it really does appear that autoimmunity as a result of X-inactivation contributes to the sex biased development of autoimmune diseases in females.
While we can’t prevent or stop X-inactivation, hopefully we can use this new knowledge to develop improved or targeted therapies for these diseases in women!
###
Dou DR et al. 2024. Xist ribonucleoproteins promote female sex-biased autoimmunity. Cell. DOI: 10.1016/j.cell.2023.12.037
Mass Spec: It's not just for chemistry nerds anymore.

It's one of the most important techniques in proteomics!
Proteomics is the study of proteins, how they function and how they interact with other molecules within our cells and tissues.
Understanding what all of those proteins are doing is important.
But not just for our own education!
Malfunctioning, missing, or improperly regulated proteins are generally what cause disease.
And quantifying what proteins are present or how they are regulated with post-translational modifications (PTMs) can tell us a lot about how they're functioning in a cell or tissue!
But, there are a number of ways we can analyze proteins.
These can include using antibodies, sequencing them directly, or detecting them with mass spectrometry (mass spec).
While the last one might sound intimidating, it’s currently the most informative of the 3!
And recent innovations have allowed these instruments to detect thousands of proteins in a single sample, including their PTMs which are important on/off switches for proteins!
Knowing that a protein is present is useful, but knowing how it is modified tells you a whole lot more about what it's doing.
Mass spec is currently the only technique capable of reliably getting both of these forms of data in a single output.
And it’s able to do this by ionizing proteins and smashing them into a detector to determine their mass-to-charge ratio or m/z.
This m/z information can then be used to figure out the original chemical composition of the things that were smashed!
There are a couple different flavors here that are worth mentioning:
LC-MS(/MS) - Liquid chromatography - tandem mass spectrometry (two detectors) is probably the simplest form of mass spec used in proteomics. LC here is used to crudely separate proteins before they’re ionized and smashed.
MALDI-TOF - Matrix assisted laser desorption/ionization - time-of-flight is a version of mass spec where the ionization of the sample is done after combining it with a ‘matrix’ that allows it to be more completely ionized. Time-of-flight (a long tube) is added to this scheme to better separate ions of different masses (the big ones take longer to hit the detector) and gives superior resolution over traditional LC-MS!
timsTOF - Adds another fun twist to time-of-flight mass spectrometry by sticking a gas tube ahead of the TOF tube to trap ions and selectively release them based on their m/z to increase resolution even further.
Orbitrap - This is something totally different although the readout is the same and it’s the highest resolution option (also the most expensive)! There’s no smashing in this setup (WTF?) but the system is able to determine the m/z of the ionized proteins as they travel in an elliptical path around a detector.
While high-throughput proteomics is still evolving, the mass spec space is one to watch closely!
Watson and Crick solved the structure of DNA and everyone lived happily ever after, right? Wrong. That was just the opening argument.
Because once the structure was proposed, everyone needed something new to fight about.
And one of those new topics was how a double stranded DNA helix was copied or 'replicated' to then be passed on to the next generation.
There were three theories for how this might work:
Conservative - A completely new copy of DNA is made that contains no pieces from the parental molecules.
Dispersive - Pieces of the parental molecule are copied and interspersed with fragments of the daughter molecules so the replicated copy is a mishmash of old and new.
Semi-conservative - One strand of a parent molecule serves as a template for creating the daughter strand so one old strand and one new strand end up in the new helix.
Watson and Crick thought the semi-conservative model was the simplest and made the most sense.
But Max Delbrück thought that was impossible because it would require the helix to totally unwind.
He believed that random copying of parent fragments was more likely so he was the champion of the dispersive model.
While the heavyweights were arguing about their theories, Matthew Meselson and Franklin Stahl, two recently minted PhDs, decided to do some actual work.
So, in 1954, they designed a series of experiments to use density gradient ultracentrifugation to settle this latest quarrel.
Now, the method they used might sound complicated but really all they did was extract DNA and spin it really really hard for a very long time - like, 45,000 rpm hard and for 20 hours.
But centrifuging the DNA was only half of the solution, they needed to figure out a way to make ‘old’ DNA strands heavy and ‘new’ ones light.
After a bit of trial and error, they settled on growing bacteria in the presence of a heavy isotope of Nitrogen, N15 and then switching the growth media to regular N14 to see what happened.
If DNA replication was semi-conservative, and not dispersive (random), they hypothesized they would see very sharp bands after centrifugation as the lighter new strands mixed with the old heavy ones.
If there was no banding, and more of a smear, they’d know replication was dispersive.
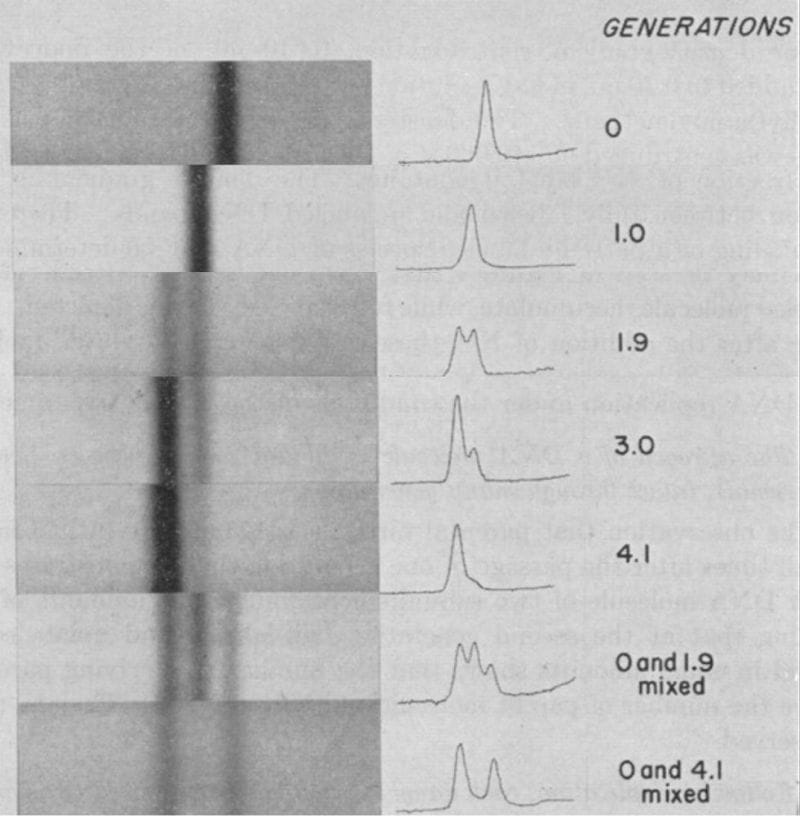
The results of this experiment can be seen above and are exactly what would be expected if DNA replicated semi-conservatively.
In Gen 0, everything is heavy, in Gen 1, the helix has one heavy and one light strand, in Gen 2, 50% of the helices are light and 50% are a mix, and by Gen 4, the majority of the DNA helices are composed of light strands.
This simple yet elegant experiment was all it took to convince Max Delbrück that he was wrong and it proved that DNA replication was semi-conservative.
###
Meselson M, Stahl FW. 1958. The replication of DNA in Escherichia coli. PNAS. DOI: 10.1073/pnas.44.7.671
