Omicly Weekly 4
December 24, 2023
Hey There!
Thanks for joining me for the Christmas Eve edition of Omic.ly!
If you're a Christmas person, hopefully you find time in between arguments with your relatives to sit back, relax and snuggle up with some Omics!
For everyone else, thanks for spending part of your Sunday reading my newsletter.
Please enjoy!
In this week's newsletter you will find:
1) The human pangenome is finally here and, in my opinion, it was the biggest story of the year
2) Why magnetic beads are the most important reagent in genomics
3) The story of how Solexa and Illumina combined forces to revolutionize the field of genomics
It's here! It's really here! The first draft of the human pangenome reference was released this year!

The Human Pangenome Reference Consortium (HPRC)'s first draft included 47 fully phased diploid assemblies of diverse individual genomes from 13 different ancestral backgrounds.
They assembled these genomes using predominantly long-read sequencing (PacBio and ONT) and genomic mapping (Hi-C and Bionano) with a dusting of Illumina Omni2.5 bead array genotyping and short-read for variant confirmation.
So why's it important that we now have 47 new, diverse, high quality genomes?
Mostly because despite sharing 99.9% of our genome with one another, there are still major differences in each of our genomes that we received from our specific ancestral lineage.
The first human genome was based mostly on a male of mixed race and did not account for all of the variation that we see across diverse populations which means it is severely lacking when it comes to helping us to determine which variants are causal of disease in different genetic backgrounds.
To fix this, the goal of the HPRC is to replace our dusty old linear reference genome with a graph genome that preserves ALL of the genetic diversity that we see across populations.
And if you don't believe that this is a big deal, let's dig in, because the benefits even with just 47 of the eventual 350 genomes are pretty impressive!
1) Variant Discovery - Showed improved performance, particularly in challenging regions and medically relevant genes - calling on average 64,000 more variants per 1kg sample and producing far fewer errors in both singletons and trios.
2) Genotyping Structural Variants - Detected significantly more SVs compared to short-read call sets, indicating that short-read SV discovery using linear reference genomes misses a significant proportion of SVs.
3) Analyzing Variable Number Tandem Repeats - VNTRs are regions in the genome that are very hard to sequence. The pangenome reduced mapping errors and enabled more accurate estimation of their length.
4) RNA-seq Mapping - Using the pangenome reduced false mapping rates, allelic bias, and increased mapped coverage on heterozygous variants, facilitating more accurate analyses of allele-specific expression.
5) ChIP-seq Analysis - Identified additional epigenetic marks that correlated well to pangenome specific structural variants with clear stratification of these marks between African and European populations!
Ultimately, the human pangenome will allow us to finally start to tease apart the complex, population-specific, genomic variants that account for all of the differences we see across populations.
While 47 genomes is a good start, I can't wait for the next 303 to be added in our quest to make genomic based healthcare more inclusive and equitable.
###
Liao WW, et al. 2023. A draft human pangenome reference. Nature. DOI: 10.1038/s41586-023-05896-x
Magnetic beads don't get much respect in the sequencing world, but they're hella useful. Here are some applications you should try!

But first, how do magnetic beads actually work?
The beads themselves are polystyrene spheres that have been coated with iron to make them superparamagnetic.
This is just a fancy term for 'they're very attracted to magnets.'
And because they're attracted to magnets, it makes them the perfect solution for high throughput isolation of biomolecules!
These iron beads can be functionalized in different ways, for example, they can be coated to bind proteins, or biotin, or even nucleic acids.
This versatility is what makes them so useful for molecular manipulation!
Nucleic Acid Extraction: Beads are functionalized with carboxyl groups which allow them to bind to salt precipitated nucleic acid. This binding is aided by a 'crowding agent' like polyethylene glycol (PEG) which forces the DNA/RNA to get up close and personal with the beads! This is basically the go-to process for anyone that wants to do high throughput nucleic acid extraction. It's also a very gentle extraction method, unlike filter based plates or columns that shear DNA/RNA, beads can yield 60kb lengths which is perfect for longer read sequencing applications.
DNA Size Selection: But wait, there's more! You can also use these beads in a similar fashion to do DNA size selection. Fragment insert size length is important for short-read sequencing and depending on your read length you might want to shoot for anywhere between 75bp to 400bp fragment sizes. Historically this was done using gel size selection or sonication, but beads can also be used to select size ranges of fragments by adjusting the concentration of the crowding agent! Essentially, the more crowding agent you include, the smaller the fragments you retain.
Nucleic Acid Normalization: Alternatively, the beads themselves have a specific binding capacity based on their size, salt concentration and PEG ratio. Because of this, these beads can be used to bind a specific amount of nucleic acid for downstream applications without having to worry about quantifying how much is there before moving on to the next step!
Target Capture: Beads can be coated with streptavidin which is a protein that binds very tightly to biotin. This affinity has been co-opted for target capture applications. Nucleic acid probes 5' labeled with biotin are incubated with a DNA sample to bind to their targets, exposed to streptavidin covered magnetic beads, and then washed to isolate the captured target from the bulk sample.
Immunoprecipitation: Yet another big word but it just means 'to use an antibody to isolate something!' Magnetic beads can be functionalized to bind antibodies. These can then magnetically isolate specific cells from a sample, proteins bound to DNA fragments, or fish out any affinity interaction you desire.
Respect the power of the bead 💪!
Illumina is considered by many to have single handedly transformed the field of genomics. They did it by building on the foundation Solexa established in 1997.
A lot of great DNA stories start in an English pub and this one isn't an exception.
Shankar Balasubramanian and David Klenerman formed Solexa after deep discussions at the Panton Arms Pub sparked a new idea for how to revolutionize DNA sequencing.
Their concept centered on the visualization of single DNA molecules as they incorporated nucleotides on a solid surface.
This sounds deceptively simple.
But, key components of this technology included the development of a reversible sequencing chemistry, a method for imaging single molecules, and a highly parallelized array with the goal of sequencing over 1 billion bases.
Sequencing single molecules proved to be a huge challenge but Solexa was able to pivot their approach by acquiring a clustering chemistry from a struggling Swiss sequencing firm named Manteia.
Their chemistry was conceptualized by Pascal Mayer and Laurent Farinelli and it created spots of ~1,000 identical molecules.
This was significant because it amplified the signal that Solexa was able to detect during each sequencing cycle.
With this foundation, they maintained a competitive edge by continuously improving on their quality.
As a result, Illumina acquired them in 2006 for $650m.
Incredibly, 2 years later, they published a full human genome sequence.
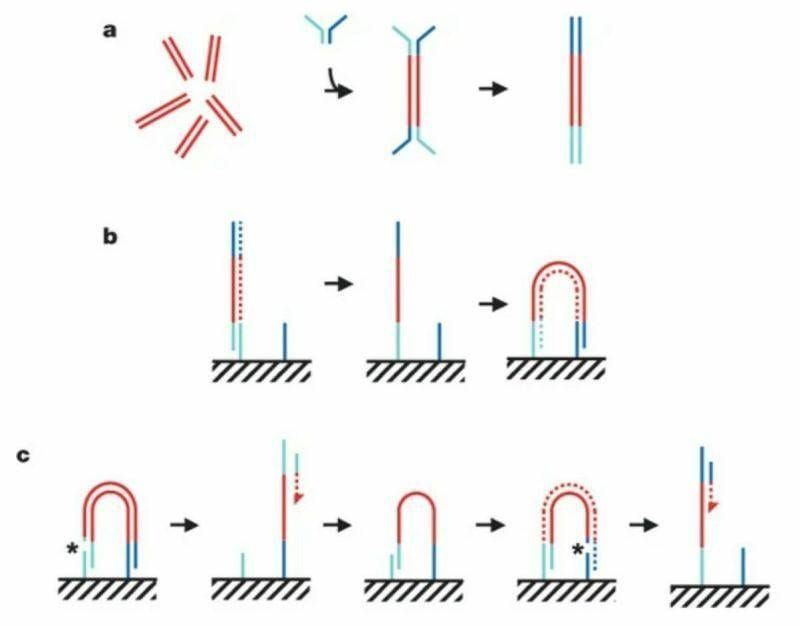
The figure above summarizes the sequencing chemistry described in that seminal paper:
Illumina-Solexa libraries are made by breaking DNA into short ~300bp fragments, ligating on adapters, and then amplifying with PCR as seen in (a).
The adapter ligation step is crucial for 2 reasons 1) it adds known sequence to an unknown fragment to amplify it with PCR but 2) that known sequence can be used to capture those molecules as seen in (b).
Capturing these molecules on a slide is critical for the process of clustering.
In (b) you can see the turquoise adapter binding to a complementary sequence on a slide, it’s copied through an extension reaction, and then 'bridges' over to another complementary sequence where the process is repeated.
This is done multiple times to create a lawn of clonally amplified clusters.
Sequencing occurs through the addition of a sequencing primer, depicted here as a turquoise bar with a red dashed arrow.
This is followed by another round of clustering and bridging to sequence the opposite end of the DNA, depicted now as the blue bar with the red dashed arrow.
In the 15 years since the publication of this paper, millions of people have been sequenced using this technology.
It's hard to imagine that the genetic revolution would have been possible without these groundbreaking innovations.
###
Bentley DR et al. 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. DOI:10.1038/nature07517
