Omicly Weekly 3
December 17, 2023
Hey There!
Thanks for joining me for the third BETA issue of Omic.ly!
I think I'm done making mistakes with the posting of these!
Well, except for the typos...they'll probably be here forever...
BUT, I've been super impressed with the open rate on the first issues:
60% Premium
50% Supporter
77% Free
Thanks so much for actually reading!
As always, send me any comments or suggestions if you think there's something I can add to improve the newsletter.
Please enjoy!
In this week's newsletter you will find:
1) A giant million sample genetic database was released in November
2) Why Transcriptomes are sometimes more useful than Whole Genomes
3) A multi-billion dollar industry was spawned from the discovery of circulating free DNA
A gigantic genomics dataset became available at the beginning of November. How gigantic?

Well, let's first put it into perspective with the other available databases:
UKBiobank: ~500,000
GnomAD: ~125,000
ExAC: ~60,000
EVS: ~6,500
1KG: ~2,500
The Regeneron Genetics Center Million Exome (RGC-ME) set is the largest of them all and includes 985,830 samples!
It more than doubles the number of samples we previously had for rare variant and population studies.
But before we get too excited, it's important to explain what an Exome is and how it differs from a genome.
The human genome is 3 billion base pairs spread across 23 chromosomes, but only about 65 million of those base pairs, or ~2%, code for protein.
This portion of the genome is referred to as the Exome and about 85% of disease causing variation occurs in the exome so while it's not all 3 billion base pairs, it covers most of the variation we care about from a genetic disease perspective.
From a broad research perspective, genomes would be way better, but a massive exome dataset like this is a goldmine for a pharmaceutical company like Regeneron who is looking for variants with interesting phenotypes that indicate potential pathways to target for therapies.
One recent example here is the discovery of the gene PCSK9 as a therapeutic target for the reduction of cholesterol. It was discovered that human 'knockouts' for PCSK9 were correlated with exceptionally low cholesterol and a potential blockbuster drug was born.
So, a million sample database can be used to find similarly useful therapeutic targets but much like our other genetic databases, this one also isn't very diverse.
For context, the demographics of the United States are:
White: 58%
Hispanic: 19%
African: 12%
Asian: 6%
Other Ethnicities: 5%
This data set is 77% white, 6% African, 5% Asian and 7% admixed American.
Despite the skew, it's still the most diverse dataset in existence just because of its sheer size.
And that means it can be used to help clean up some of our blindspots!
The researchers discovered that fewer pathogenic variants were found in non-whites but that non-whites also had more variants of unknown significance with Africans having the greatest number of variants in their genomes when compared to the standard reference.
These results highlight the need for both reference diversity and including diverse populations in studies so that pathogenic variation can be identified across all ethnicities.
To this end, RGC-ME was released in aggregate as a part of a public variant browser similar to ExAC/GnomAD with fine breakdowns of variant allele frequencies by population.
This is huge because RGC-ME includes, "10.5 million missense (54% novel) and 1.1 million predicted loss-of-function variants (65% novel, 53% observed only once)."
###
Sun KY et al. A deep catalog of protein-coding variation in 985,830 individuals. DOI: 10.1101/2023.05.09.539329
Transcriptomes: underappreciated, underutilized, and sometimes more useful than WGS.

The transcriptome is usually considered a lesser -ome and it’s often overlooked when we talk about rare disease diagnosis, but it shouldn’t be.
Transcriptomics is the study of the all of the RNA sequences that are present within a cell.
The transcriptome is a product of our genome and differences in RNA expression can regulate what proteins are ultimately made.
This determines the functions performed by our tissues which themselves are made up of trillions of cells, all with slightly different transcriptomes.
So for us to really, actually, understand anything about what our genome is doing at any given time, we need to look at the functional output of the genome.
This is especially important in the context of whole genome sequencing because it will find about 5 million variants, the vast majority of those having unknown effects.
And in some of the tougher to solve cases, where a deep intronic or non-coding variant results in disease, we can use transcriptome data to make sense of everything.
Transcriptomes give us 3 important read-outs:
A quantitative measure of gene expression - How much of each RNA is present? Anything missing? More than we expect?
The sequence of expressed transcripts - This is a bit like a double-check of genome sequencing. Do we find differences between what was in the DNA sequence and what is expressed in the RNA? Fusions?
Which isoforms are present - So the genome codes for like 22,000 genes but each of those genes can be alternatively spliced to create more than one version of a protein. 93% of genes undergo this process, so you can see how this can get complicated quickly. And if mutations affect splicing, we’ll see that in what bits of the genome actually make it into the RNA!
The biggest drawback of transcriptomes is that sometimes tissue specific collections are required, especially for muscular disorders, since the cells in our body don't all express the same RNAs.
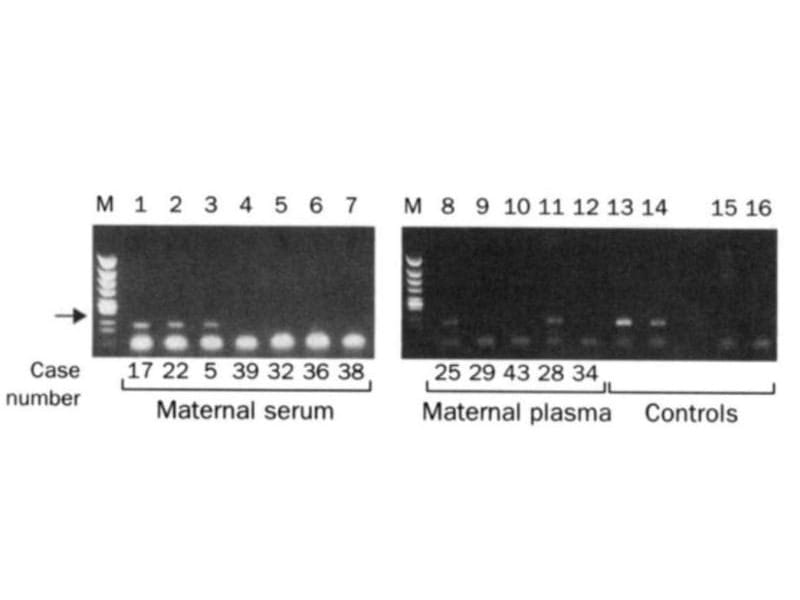
The two gels below spawned a multi-billion dollar industry that didn't exist prior to their publication in 1997.
While it may seem obvious today, you might find it surprising that we didn't know that fetal DNA was present in a pregnant mother's bloodstream until the late 1990's.
Prior to this discovery, genetic testing on fetuses was only performed if a problem was suspected with a pregnancy or if there was a family history of genetic disorders.
This testing was performed by karyotyping fetal cells.
You've probably seen one of these 'chromosome spreads' in a biology textbook where each of the 23 pairs of chromosomes are lined up next to one another.
This allows a geneticist to check that the chromosomes are intact and see if there are any abnormalities such as those found in Down Syndrome where patients have 3 copies of chromosome 21 instead of 2.
But, back in 1997, the process for collecting fetal cells was very invasive and was done using one of two techniques:
Amniocentesis (Amnio) - a 3-5" long needle is inserted into the mother's abdomen to collect 20 milliliters of amniotic fluid for testing.
Chorionic Villus Sampling (CVS) - A 6" long needle is guided via ultrasound, either vaginally or through the abdomen, to obtain tissue from the placenta for testing.
Unfortunately, these procedures carry a risk and 1-2% of the time they can lead to the loss of the fetus.
Recognizing that there had to be a better way, Yuk-Ming Dennis Lo set about figuring out how to get at fetal DNA without having to perform Amnio or CVS.
He knew that fetal cells made it into the maternal bloodstream and that mother and child exchanged cellular material.
But he couldn't isolate enough fetal cells from the blood to do prenatal genetics with them.
Luckily, in 1996, Lo heard that a team in Switzerland had shown that tumors actually shed cell free DNA into the bloodstream and this tumor DNA could be detected using PCR and primers specific to the tumor DNA.
He reasoned that a baby, or more accurately, the placenta, was basically like a giant tumor and shared the bloodstream with the mother.
So, it made sense that it too should shed cell free DNA into the bloodstream.
The figure above is proof that Lo's hypothesis was correct and fetal DNA could be isolated and amplified from the blood of pregnant females.
In this figure, the arrow highlights a 198bp PCR product from the Y chromosome. Case numbers greater than 30 are pregnancies with female fetuses (don't have a Y), and case numbers less than 30 are male fetuses (have a Y).
Further technical advancements and the invention of high throughput sequencers have transformed this discovery into a popular pregnancy screening test.
Today, non-invasive prenatal testing is second only to oncology testing in sequencing market share.
###
Lo YMD, et al. 1997. Presence of fetal DNA in maternal plasma and serum. The Lancet. DOI:10.1016/s0140-6736(97)02174-0
