Omic.ly Weekly 64
February 24, 2025
Hey There!
Thanks for spending part of your week with Omic.ly!
This Week's Headlines
1) AI is making new, functional, enzymes now
2) Magnetic beads might be the most useful reagent in genomics
3) Early pregnancy tests were wild
Here's what you missed in this week's Premium Edition:
HOT TAKE: Roche's short-read nanopore sequencer is going to be entering a very crowded marketplace
Or if you already have a premium sub:
Artificial intelligence has been used to develop a new enzyme that can catalyze a multistep chemical reaction!
Enzymes are like little chemistry magicians.
They make complex chemical reactions happen at room temperature that would be nearly impossible to perform outside of an advanced laboratory.
And they’re able to perform these amazing chemical feats because enzymes work by grabbing onto molecules and forcing them together (or pulling them apart) at the molecular level.
All of this happens within the “active site” of an enzyme which is a cavity within the protein that interacts with the molecules that are participating in the chemical reaction.
But enzymes don’t JUST force molecules together, sometimes they become intermediaries within the process and covalently bond to these molecules to get them into the right state to perform the next step in the reaction.
That is to say, this is all really complex and it’s a wonder that enzymes have evolved to be able to perform these fantastic feats.
It also means that designing our own enzymes is incredibly tough, and until recently our best techniques for building new enzymes involved processes like “directed evolution” or just plain old trial and error.
Directed evolution is where you take an organism (usually a bacterium), expose it to nasty conditions containing the molecule you want it to deal with, and through multiple rounds of selection, find a bacterium that eventually develops useful enzymes that do the thing you want them to do.
But this is a time consuming process and rarely ends up producing enzymes that are highly efficient!
There has to be a better way, right?!
That better way for a long time seemed like it was going to involve computational biology, but even using complex supercomputers, we couldn’t really develop useful enzymes that could perform multi-step chemical reactions!
Usually what got made could perform single steps, but then they would be gummed up in the process before moving on to subsequent steps.
That’s super frustrating!
However, we now have AI algorithms that we can employ to help us iterate through candidate protein that might be able to help us produce enzymatic magicians of our own.
In this week’s paper, researchers attempted to use AI to develop novel serine hydrolases.
These are enzymes that catalyze the breaking of ester bonds using water!
While that sounds simple, breaking an ester bond using an enzyme is a 3 step reaction!
And previous attempts at making this type of ester hydrolase from scratch have ended in failure.
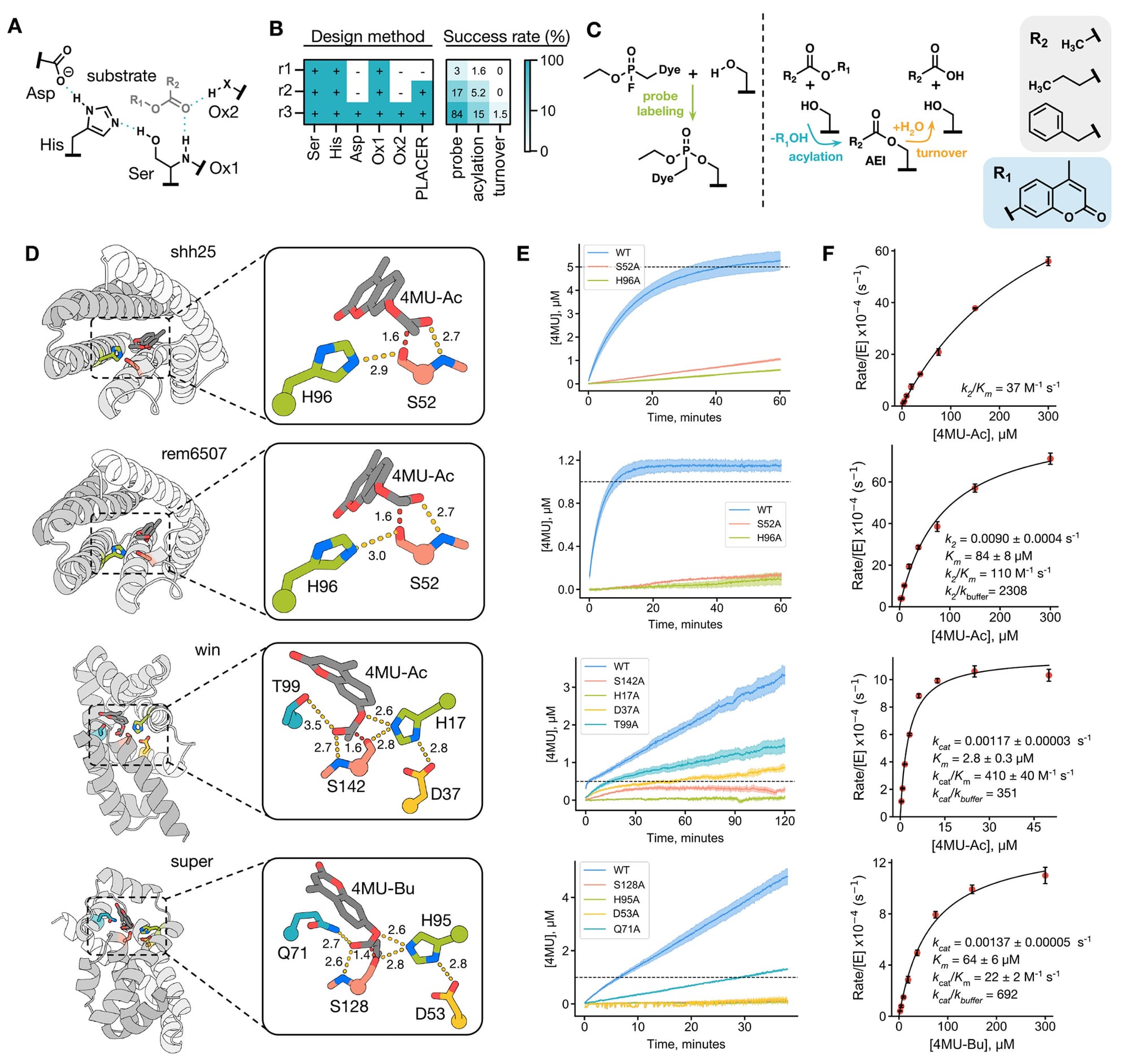
In the figure above, though, you can see that this diffusion based AI ended up finding a few COMPLETELY NEW proteins that actually worked!
A is a model of the active site, B has a table showing what parts of the active site were designed during each round (r1, r2, r3) and then a success table to the right of that showing that only 1.5% of tested enzymes could complete the multi-step (turnover) reaction!, C shows the 3 phases of the reaction and substrates, D shows 4 enzyme structures with a zoom in of their active sites (folded in alphafold 2), E shows the activity of these 4 enzymes - The first two were made in design round 1 and have a plateau which means they didn’t make it past the first step of the reaction while the bottom two enzymes don't show a plateau because they could complete the entire reaction cycle (these are the 2 enzymes that worked in design round 3!), and F is a Michaelis-Menten plot of the catalytic activity of each enzyme.
The researchers went on to show that they could further improve the catalytic activity of the round 3 enzymes and they were able to create at least one that had similar activity as a naturally occurring serine hydrolase.
But the big step forward here is that these newly designed hydrolases have completely different folded structures than their biological counterparts meaning that AI could soon aid in the development of totally novel proteins.
One thing to keep in mind here though is that this AI didn’t just spit out immediately useful structures!
There was a lot of screening that needed to be done here.
Each design round ended with ~200 candidates that were expressed in e. Coli and then tested for their catalytic activity before being used to seed the next design round.
This exquisitely highlights the need for validating the outputs of the solutions that come out of any generative AI model.
###
Lauko A, et al. 2025. Computational design of serine hydrolases. Science. DOI: 10.1126/science.adu2454
Magnetic beads don't get much respect in the sequencing world, but they're hella useful. Here are some applications you should try!

But first, how do magnetic beads actually work?
The beads themselves are polystyrene spheres that have been coated with iron to make them superparamagnetic.
This is just a fancy term for 'they're very attracted to magnets.'
And because they're attracted to magnets, it makes them the perfect solution for high throughput isolation of biomolecules!
These iron beads can be functionalized in different ways, for example, they can be coated to bind proteins, or biotin, or even nucleic acids.
This versatility is what makes them so useful for molecular manipulation!
Nucleic Acid Extraction: Beads are functionalized with carboxyl groups which allow them to bind to salt precipitated nucleic acid. This binding is aided by a 'crowding agent' like polyethylene glycol (PEG) which forces the DNA/RNA to get up close and personal with the beads! This is basically the go-to process for anyone that wants to do high throughput nucleic acid extraction. It's also a very gentle extraction method, unlike filter based plates or columns that shear DNA/RNA, beads can yield 60kb lengths which is perfect for longer read sequencing applications.
DNA Size Selection: But wait, there's more! You can also use these beads in a similar fashion to do DNA size selection. Fragment insert size length is important for short-read sequencing and depending on your read length you might want to shoot for anywhere between 75bp to 400bp fragment sizes. Historically this was done using gel size selection or sonication, but beads can also be used to select size ranges of fragments by adjusting the concentration of the crowding agent! Essentially, the more crowding agent you include, the smaller the fragments you retain.
Nucleic Acid Normalization: Alternatively, the beads themselves have a specific binding capacity based on their size, salt concentration and PEG ratio. Because of this, these beads can be used to bind a specific amount of nucleic acid for downstream applications without having to worry about quantifying how much is there before moving on to the next step!
Target Capture: Beads can be coated with streptavidin which is a protein that binds very tightly to biotin. This affinity has been co-opted for target capture applications. Nucleic acid probes 5' labeled with biotin are incubated with a DNA sample to bind to their targets, exposed to streptavidin covered magnetic beads, and then washed to isolate the captured target from the bulk sample.
Immunoprecipitation: Yet another big word but it just means 'to use an antibody to isolate something!' Magnetic beads can be functionalized to bind antibodies. These can then magnetically isolate specific cells from a sample, proteins bound to DNA fragments, or fish out any affinity interaction you desire.
Respect the power of the bead 💪!
For the better part of 40 years, pregnancy tests were done by injecting urine into animals and seeing what happened. No, seriously!
Prior to 1928, determining whether someone was pregnant was a waiting game.
However, that didn’t stop people from trying to predict whether a missed period meant that a baby was on the way.
These early 'tests' ranged from mundane to insane and usually involved 'analyzing' urine.
The reason for this is probably due to its proximity to the birth canal.
And purveyors of these early tests were often referred to as 'piss prophets.'
This hints at the accuracy of such methods, but urine was actually the ideal sample type!
Before we get to WHY that is, we need to talk about the birds and the bees.
We know now that one of the first hormones to appear after sperm fertilizes an egg is human chorionic gonadotropin (hCG).
It is produced by the trophoblast cells that surround a developing embryo which eventually go on to form the placenta.
hCG’s function is to signal the ovaries, specifically the corpus luteum, to continue producing progesterone and estrogen.
This prevents the sloughing off of the endometrium and its discharge which marks the end of the menstrual cycle.
It’s these hormones that serve as the basis for modern pregnancy testing.
But in the early 1900’s, ‘hormones’ were a totally new concept with the word itself being coined by Ernst Starling in 1905.
By the 1920’s, scientists were finally starting to unravel the secrets of how hormones regulate the differences between the biological sexes.
Early work by Selmar Aschheim and Bernhard Zondek showed that extracts from the anteropituitary (the back of the pituitary gland) could promote ovary development and growth in animal models.
In 1927, they discovered that the urine of pregnant mice contained many of these same hormones and it had a similar stimulatory effect on their ovaries.
They quickly realized that injecting human urine into mice and seeing if their ovaries grew could be used to diagnose pregnancy and published on this in 1928.
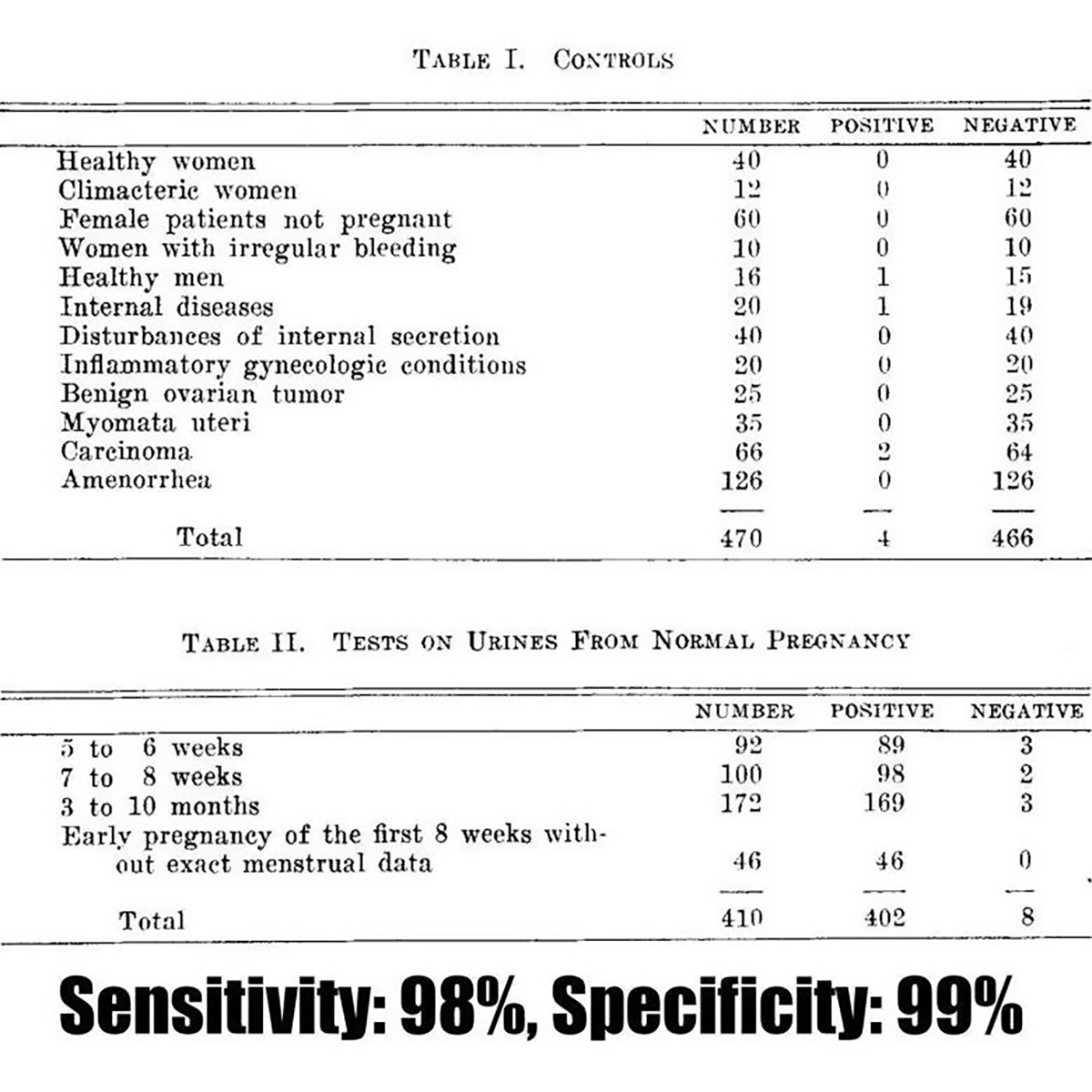
Unfortunately, this paper has no figures, but the figure below is from a paper published in 1930 detailing the utility of the Ascheim-Zondek test for the diagnosis of pregnancy and placental tumors.
The two tables above display the impressive performance of the test in detecting 98% of pregnancies!
Further improvements came in 1936 with the Friedman or 'Rabbit' test in the US that gave results in 24hr, along with the development of a version using frogs that was popularized in the UK.
Animal based testing finally stopped in the 1960's when Leif Wide and Carl Gemzellthe invented an immunoassay to detect hCG directly.
However, the biggest innovation came in 1978 when at-home pregnancy tests first became available over the counter.
###
Aschheim S. 1930. The early diagnosis of pregnancy ... by the Aschheim-Zondek test. Amer J Ob Gyn. DOI:10.1016/s0002-9378(30)90238-4
Were you forwarded this newsletter?
LOVE IT.
If you liked what you read, consider signing up for your own subscription here:
