Omic.ly Weekly 61
February 3, 2025
Hey There!
Thanks for spending part of your week with Omic.ly!
This Week's Headlines
1) SNPs better watch their back, a new variant class is looking to steal the MRD limelight
2) The $100 genome is a myth
3) The isolation of insulin required a lot of trial and error
Here's what you missed in this week's Premium Edition:
HOT TAKE: Roche finally has something to say about what they've been doing for the past 10 years with Genia and Stratos Genomics
Or if you already have a premium sub:
Watch out, Structural Variants are coming to MRD testing!
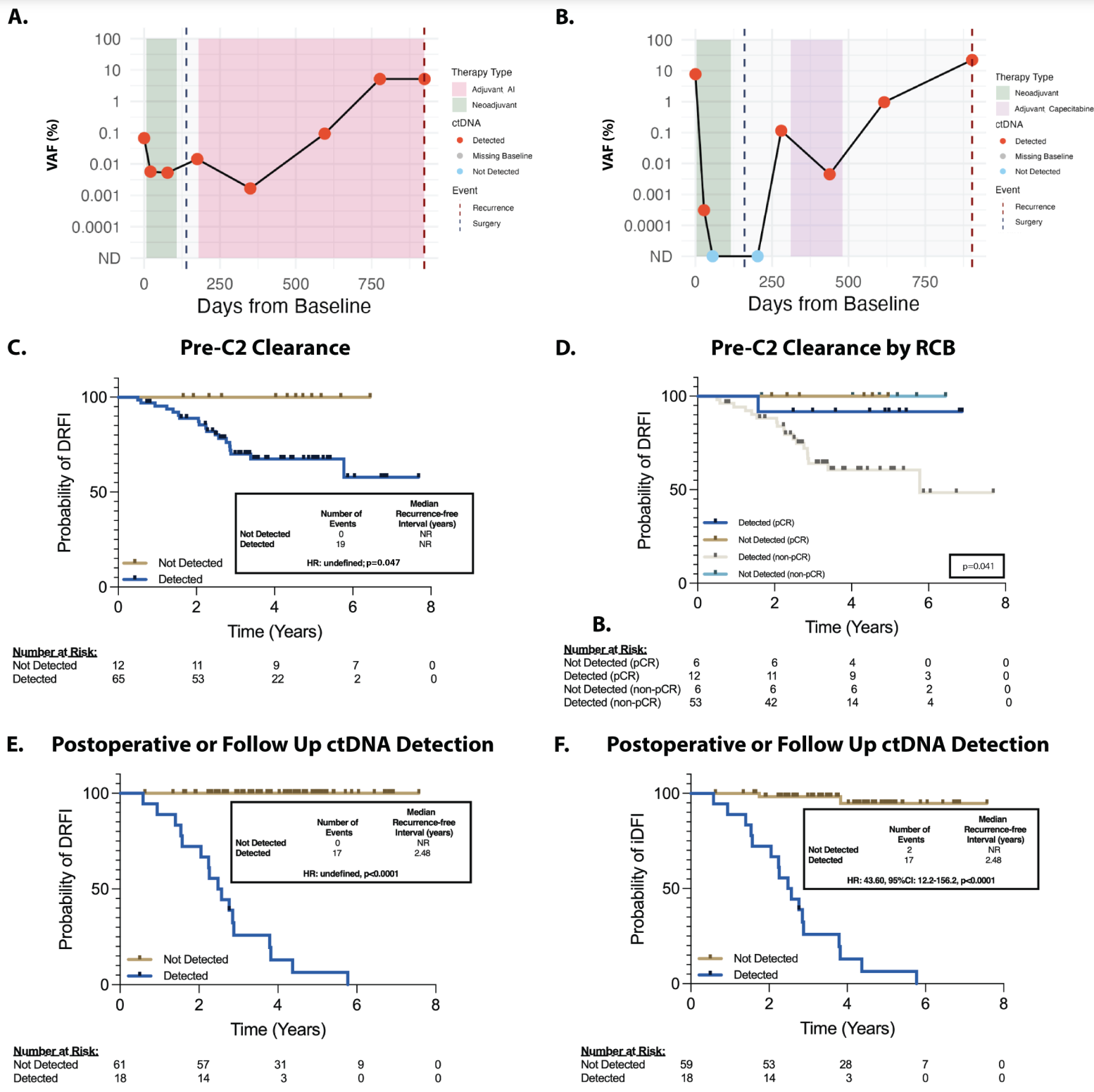
The goal of minimal residual disease (MRD) testing is to determine how much disease remains in cancer patients after a solid tumor is removed or a hematologic (blood) cancer is treated.
In hematologic cancers, MRD tests using PCR were first seen in the late 1980’s.
These tests targeted the IG-TR gene rearrangements that were common in lymphoid malignancies.
PCR allowed oncologists to ditch using Southern blots to detect these changes and improvements in MRD tests led to quantitative PCR based versions in the late 90’s.
But, the challenge with using PCR for this sort of testing is you need to know what you’re looking for to detect it!
If you don’t have, or know of, a common mutation in a cancer, it’s very hard to determine the residual disease burden, or detect when a cancer comes back after treatment.
Fortunately, the advent of massively parallel sequencing in the early 2000’s gave us a new tool for tracking cancer progression and finding unique markers of disease.
But the use of high throughput sequencing as a diagnostic in oncology has been prohibitively expensive until more recently.
The advent of tumor-informed high-throughput sequencing based MRD assays really began in 2018 with Natera’s publication of their Signatera product which is a single-nucleotide polymorphism (SNP) based test.
It uses a patients’ own tumor tissue to develop a SNP fingerprint that is then assayed longitudinally (over months/years) after cancer removal to see if that DNA signature reappears in a patient’s blood sample.
Reappearance of that signal is indicative of cancer’s return, and the hope is that these MRD tests can detect this recurrence in time for us to do something about it!
But SNPs aren’t the only kinds of mutations that turn up in cancers!
They’re also full of large structural variants (SV) like gene deletions, duplications and other large genomic rearrangements.
The researchers behind today’s paper showed that these SVs can also be used to generate MRD fingerprints that perform as well as or better than their SNP based counterparts!
To do this they 15x whole genome sequenced tumor tissue from breast cancer patients to identify tumor specific structural variants and created a 16 marker fingerprint.
But instead of using that fingerprint to do MORE sequencing, they instead used an ingenious method to develop SV breakpoint specific digital PCR (dPCR) assays.
Using dPCR allows them to detect single molecules in blood without having to spend a bunch of money sequencing stuff they don’t care about!
And in the figure above you can see that their hard work paid off! A and B) Show representative plots of ctDNA detection before surgery (green area), at surgery (black dashed line), after surgery and during chemo (pink and purple) and at detection of disease recurrence (red dashed line) C and D are kaplan-meier curves (track the death of subjects in a trial) showing that the detection of ctDNA in the neoadjuvant setting (before tumor was removed) was associated with poorer prognosis and that the same was true in the adjuvant setting (after the tumor was removed).
This test detected 100% of cancer recurrence with a median lead time in early breast cancer of 417 days.
That’s 417 more days to try to figure out how to delay the cancer a second time!
###
Elliot MJ, et al. 2025. Ultrasensitive Detection and Monitoring of Circulating Tumor DNA using Structural Variants in Early-Stage Breast Cancer. DOI: 10.1158/1078-0432.CCR-24-3472
Disclosure: I have received consulting fees from SAGA Diagnostics (their Pathlight assay was used in this paper)
The $100 genome is a myth, let's focus on the $0 one instead.

One of the biggest disappointments in the 'cost of a genome' discussion is that despite reducing costs, access to genome sequencing for disease diagnosis is very hard to come by outside of an academic medical center.
First it was the $1,000 genome, then we got a $500 genome on the NovaSeq, now it's ~$200 on the Xplus and Aviti with a ~$100 genome with Ultima and Complete.
The problem is these numbers mean basically nothing to the vast majority of patients waiting to get a diagnosis.
Why?
Well, for one, that number doesn't include the cost of labor, analysis, or interpretation.
In the best case, for a clinical workup of a singleton (the affected individual), we're looking at $50 for sample collection, DNA extraction, library prep, and labor; $100-200 for sequencing; and then between $300 and many thousands of dollars for analysis/interpretation.
If you're doing trios (Mom, Dad, affected individual), you can basically triple those costs.
So even in the best case scenario, we're talking ~ $400, and that's just Cost of Goods Sold (COGS) without adding in any profit.
If you're doing this with a major non-profit, it's ~$9,000 for a trio, but that includes case review by a world class clinical team.
It's likely that equivalent quality can be had for much less, and the future integration of AI and other emerging methods should make these prices more palatable to insurance companies.
And that's what I'm most excited about:
The $0 genome.
Because these $9,000 clinical workups are generating the evidence needed to get insurance companies to pay for them.
My hope is that the majority of payers will finally wake up and say, "Maybe it doesn't make sense for these kids and families to wait a decade for a diagnosis?"
We're starting to see some of this already in more progressive states where they're funding rapid neonatal sequencing programs for very sick kids - California, Michigan, Oregon, Maryland, Minnesota, and others.
But many of these programs have pre-authorization requirements that restrict access to this much needed testing.
Even UnitedHealthcare has started covering sequencing but only when a genetic cause is suspected and presentation includes any of the following: "multiple congenital anomalies (must affect different organ systems); moderate, severe, or profound intellectual disability diagnosed by 18 years of age; or global developmental delay, or epileptic encephalopathy with onset before 3 years of age."
This is progress but coverage restrictions like these mean a major swath of the population that are affected by late onset genetic disease, and who could benefit from a genetic diagnosis, will be missed.
Unfortunately, there's still work to do, and that includes expanding coverage to carrier, inherited disease, and risk factor screening in healthy people.
Baby steps, I guess.
Insulin: It was believed to exist, but everyone who tried to isolate it failed.
It was finally captured in 1921.
Many decades worth of medical observation and dissection showed that problems with the pancreas caused diabetes.
The classical symptoms are high blood sugar, glycosuria (excretion of sugar in the urine), ketosis (burning fat instead of sugar), seizures, and, if untreated, death.
It was discovered that animals who suffer from diabetes naturally lose their pancreatic islet cells, and in 1909, Jean de Meyer, and in 1913, Edward Sharpey–Schäfer, suggested that a hormone was secreted from the islet cells to control glucose metabolism and its uptake from the bloodstream.
Sharpey–Schäfer named this substance 'insuline.'
But, this is not the only function of the pancreas.
It also produces trypsin, an enzyme that helps digest proteins in the stomach.
This made things complicated because everyone who tried to isolate insulin ultimately failed because it was immediately chopped up during extraction by trypsin!
Fortunately, Frederick Banting, a Canadian surgeon, came across an article describing how the blockage of the pancreatic ducts in a patient resulted in the death of the acinar cells (the ones that pump out trypsin), but preserved the islets with no sign of the development of diabetes.
Banting, who had no formal scientific training, thought that a good way to isolate insulin might be to 'ligate' or tie off the pancreatic ducts to kill all the acinar cells and then grind up what was left over.
He pitched his grand idea to JJR Macleod, a professor of physiology at the University of Toronto, who had a lab and access to research animals.
Macleod was skeptical, but he was also going on vacation, so he gave Banting the keys to the lab, 10 dogs, and a coin flip determined that Charles Best would help Banting.
Best was also tasked with making sure the lab didn't burn down while Macleod was away.
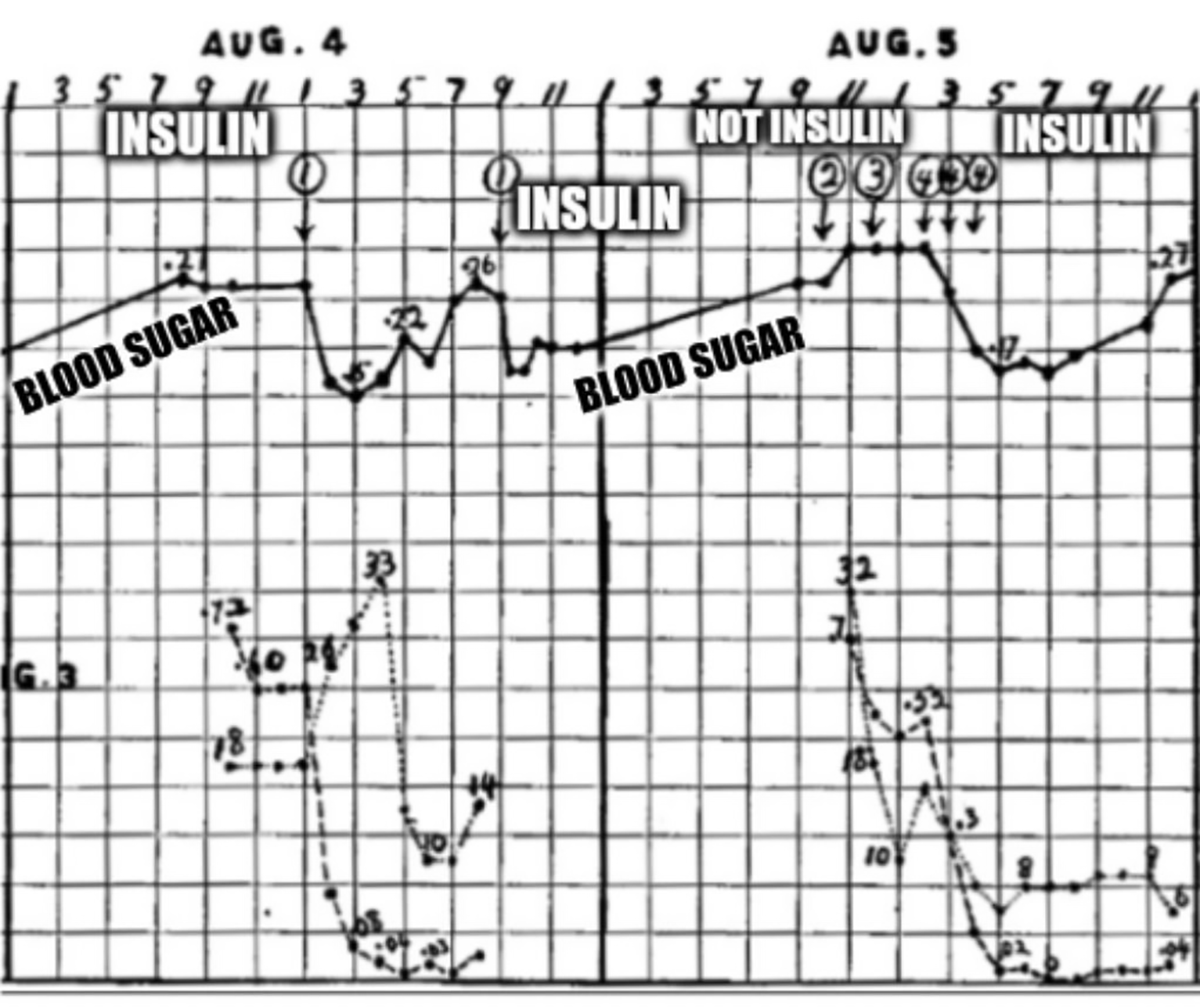
7 dead dogs later, Banting and Best had perfected their technique, and generated the data that can be seen in the figure above.
What's displayed is two days worth of blood glucose readings (solid black line) from a dog whose pancreas was removed (causes diabetes).
The numbered circles are injections of ground up organ extracts: 1. Refrigerated ligated pancreas, 2. Liver, 3. Spleen, and 4. Fresh ligated pancreas.
Here you can see that ligated pancreatic extracts (1 and 4) cause a sharp reduction in blood glucose.
Insulin had finally been isolated!
Further work by James Collip revealed that highly purified insulin could be made from chilled pancreas (no ligation) and extraction in 90% ethanol.
This process was optimized by Eli Lilly for use on pig and cow pancreas which produced the majority of insulin until a synthetic was developed 56 years later in 1978.
###
Banting FG, Best CH. 1922. The internal secretion of the pancreas. J Lab Clin Med. DOI: 10.1111/j.1753-4887.1987.tb07442.x
Were you forwarded this newsletter?
LOVE IT.
If you liked what you read, consider signing up for your own subscription here:
