Omic.ly Weekly 59
January 20, 2025
Hey There!
Thanks for spending part of your week with Omic.ly!
This Week's Headlines
1) Listen up! Fish gills are more important than you might think
2) If you thought enhancers and promoters were easy, think again!
3) The weird color patterns of corn kernels led to a major genetic discovery in 1950
Here's what you missed in this week's Premium Edition:
HOT TAKE: Everything Omicly that happened at the 2025 JP Morgan Healthcare Conference
Or if you already have a premium sub:
Did you hear? Your ears evolved from fish gills!
One of the defining characteristics of mammals are their prominent external ears.
Dogs have them.
Cats have them.
Bats have them.
You have them.
But up until now we didn’t really have any idea how mammalian ears came into being.
Physical structures usually evolve from other things - like fish fins turning into mammalian arms over eons.
However, since ears are made of non-bony cartilage, it’s been impossible to see where external ears came from in the fossil record!
There’s been some speculation that much of the programming for mammalian ears was derived from fish and that fish spiracles (help some fish breathe and are situated behind their eyes) provided the instructions for our inner ears.
But how our external ears sprung forth from our heads, and what fish parts might have aided in that outgrowth has remained a mystery.
That all changed last week with the publication of a paper showing that fish gill cartilage expresses the same genes as human outer ear cartilage.
You might be thinking that this was as simple as isolating ear and gill tissue and seeing which genes each expressed the most.
But gene expression is tricky, and sometimes the most expressed things in a tissue aren’t the most important for their structural development!
Much of the time, particularly when it comes to developmental biology, it’s transcriptional regulation of what gets expressed (and when) that drives the structural development of a tissue.
And with respect to mammalian ears, it looks like we co-opted a fish gill transcriptional regulatory network to do it!
To figure this out, the researchers behind today’s paper decided to look not just at what genes were expressed in ears and gills, but also how that expression was regulated throughout development.
They did this using single-cell RNAseq (gene expression) and ATACseq (chromatin accessibility - ie finds enhancers and active regions) to compare differentially accessible regions (DARs) in the genome.
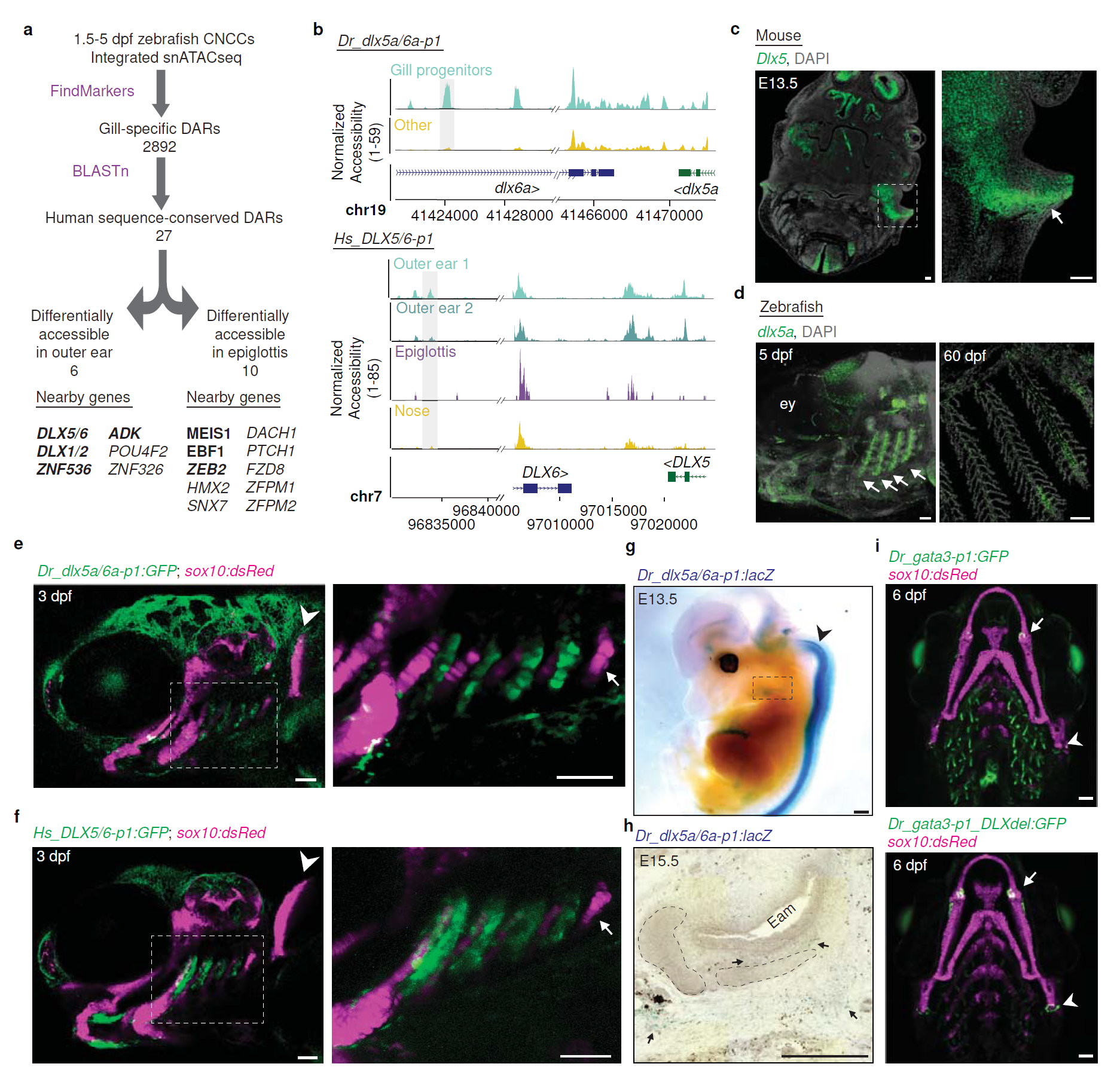
What they found can be seen in the figure above a) schematic of how they identified regions and genes of interest in fish gills and mammalian outer ears - some of the top hits were developmentally important transcription factors (bind to DNA and help turn it into RNA) like DLX1,2,5,and 6
b) DLX5/6 genome accessibility plots comparing fish (top) to humans (bottom) and show that the gill and ear accessibility are quite similar
c, d) RNA in situ hybridization (shows you where the RNA is) of DLX5 in the mouse head (arrow points to the developing outer ear) (c) and fish gills (arrows point to developing gills) (d)
e,f) Human DLX5/6 (f) is expressed in the same places as fish DLX5a/6a (e) when its put into a fish embryo
g,h) and the same is true when you put fish DLX5a/6a into a mouse!
i) is a control showing that when you delete the DLX5/6 DAR, you lose gill expression (green).
Interestingly, mice who have deletions of DLX5 and 6 also lack outer ears, but this is the first time that this gill-ear transcriptional connection has been made!
###
Thiruppathy M, et al. 2024. Repurposing of a gill gene regulatory program for outer ear evolution. Nature. DOI: 10.1038/s41586-024-08577-5
And now for something (not so) straightforward: Enhancers and Promoters

One of the most basic concepts we learn in Biology 101 is that enhancers and promoters are responsible for turning DNA into RNA.
This is called DNA transcription.
At the most basic level, it requires the recruitment of RNA polymerase to DNA.
Luckily, RNA polymerases are attracted to a specific sequence in DNA:
The TATA box!
This is a sequence found within the ‘core promoter’ and the most common ‘consensus’ sequence is TATA(A/T)A(A/T).
It is recognized and bound by TATA Binding Protein (TBP).
And TBP is what actually attracts the polymerase to the promoter.
Once the polymerase sits down, transcription is ‘initiated’ and ultimately leads to the production of RNA.
That RNA is then translated into protein.
However, sometimes the promoter is hard to find, and this is where enhancers come in!
Enhancers are sequences that work at a distance (not really, but we’ll get to that) and attract a dog pile of other proteins called transcription factors.
These transcription factor bound enhancers act as little light houses that scream, “Hey, over here, dummy!”
And this allows the polymerase to find the promoter and start transcription!
But, what if I told you that 85% of human promoters don’t have a TATA box?
Unfortunately, the advent of genome-wide techniques showed us that things aren’t so simple!
Using techniques like Chromatin Immunoprecipitation (ChIP) we pulled RNA polymerases and their transcription factor friends out of solution with antibodies and sequenced the DNA that they were stuck to.
85% of the time, those sequences DIDN’T include a TATA-box.
Wild.
We also learned that enhancers actually don’t act at a distance!
They are brought very close to the promoters they assist through the actions of proteins that modify the structure of our chromosomes!
We figured this out through another genome-wide method, Chromosome Conformation Capture, that allowed us to look at what parts of chromosomes touched one another.
That, combined with ChIP, showed us that chromosomes form ‘loops,’ and those loops often work together with transcription factors to bring enhancers and promoters together to initiate transcription.
But wait, there’s more!
We also discovered that transcription is quite promiscuous.
The simple Biology 101 description says that RNA polymerase turns DNA into RNA for the sole purpose of translating it into protein.
But, as we’ve learned, NOTHING in biology is ever simple.
RNA polymerases actually transcribe whatever they can sit on.
And within this enhancer-promoter (E-P) complex, there’s a lot of DNA to sit on!
So, we get RNA from the DNA around enhancers (eRNA).
And also RNA from the complementary DNA strand, (upstream antisense RNA - uaRNA).
The purpose of these RNAs is debatable but it’s thought that all of this activity prevents the E-P complexes from getting turned off.
See, like I said, it's (not so) straightforward!
While everyone else was distracted by the structure of DNA, Barbara McClintock was discovering a little thing called the transposable element.
The 1950's were a turning point in the fields of molecular biology and genetics.
For many years, scientists studied genetics but weren't entirely sure how it all worked on the molecular level.
They were aware of chromosomes, knew that they were composed of DNA and protein, but there was still a big question about the functional role of those two components in inheritance.
Fortunately for McClintock, who studied genetics in maize (corn), it didn't matter much what the genetic material was but that it could be tracked and its effects seen in the offspring of her crosses.
As an early cytogeneticist (scientist that studies chromosomes), McClintock developed many of the foundational techniques that were required for studying chromosomes in maize and she produced the first genetic maps of all 10 chromosomes.
Her early work was focused on studying inheritance and she was one of the first to observe and describe the phenomenon of homologous recombination, or crossing-over, which is when two chromosomes exchange genetic information.
She published this work in 1931 and continued to study maize genetics until her curious observation that pieces of chromosome 9 appeared to move around the genome.
But these pieces, Ac (activator) and Ds (inhibits pigment production), didn't just move around, Ac seemed to control Ds, and ultimately the color patterns of the maize kernels!
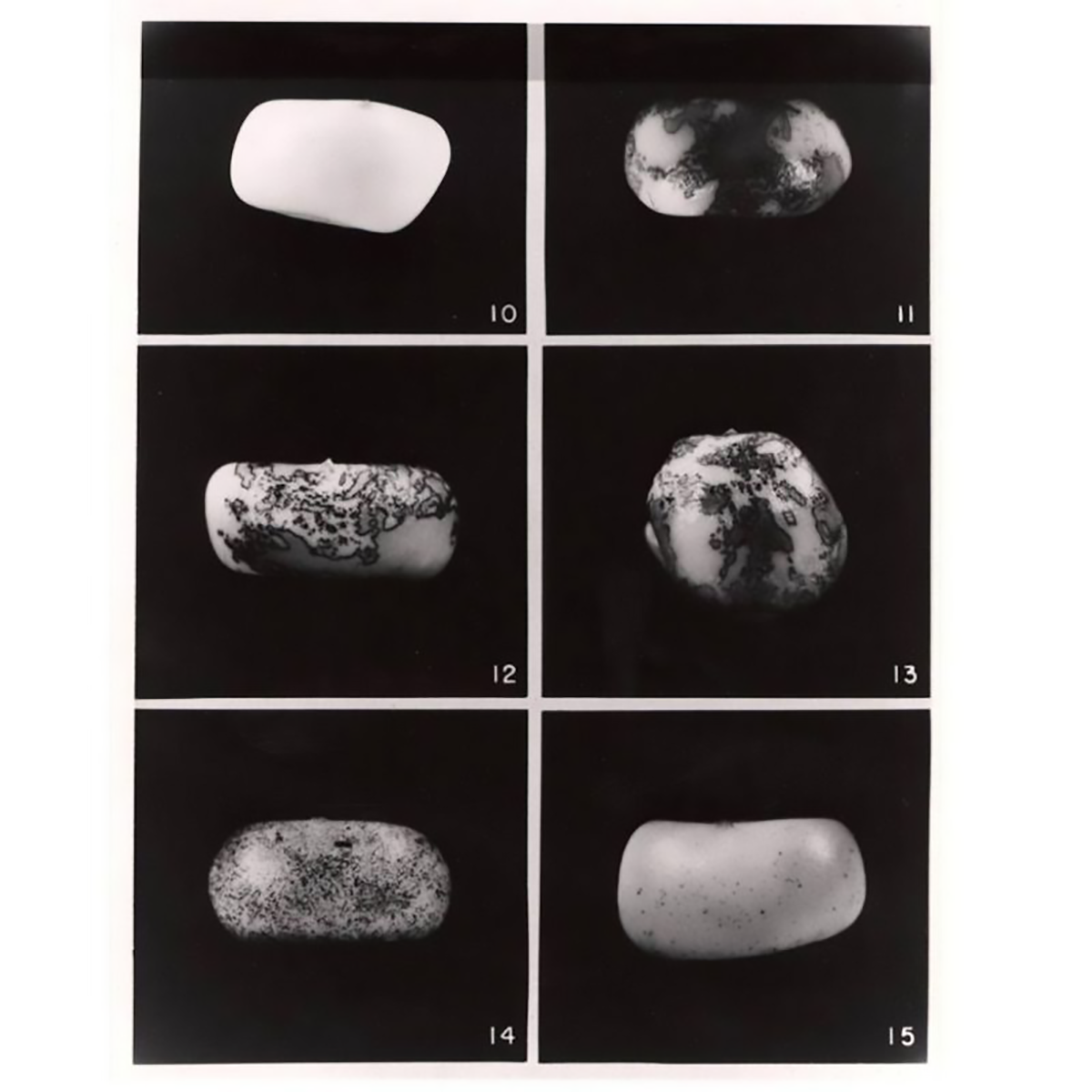
The figure above is an image of the kernels that McClintock described in, 'The origin and behavior of mutable loci in maize.'
Kernel 10 - no Ac, 11-13 - 1 copy of Ac, 14 - 2 copies, 15 - 3 copies.
We know today that 66% of the human genome and 85% of the maize genome is made up of these mobile 'transposable' elements and they are key in our understanding of epigenetics, but back in 1950, this discovery was problematic.
At the time, it was thought that genomes were static and ordered; pieces of them did not move around.
McClintock's findings were so poorly received that she stopped formally publishing them after 1953 because she thought her colleagues just weren't interested.
Fortunately, her work was 'rediscovered,' or, more accurately, 'understood,' in the 1970's when similar elements were found in bacteria, flies and humans.
She was awarded the Nobel in 1983 for the discovery of transposable elements, which was 3 decades after her original description of them.
McClintock's story in science isn't dissimilar from that of other prominent women, like Rosalind Franklin.
However, unlike Franklin, McClintock lived long enough to finally be properly recognized for her historic achievements.
###
McClintock, B. 1950. The origin and behavior of mutable loci in maize. PNAS. DOI: 10.1073/pnas.36.6.344
Were you forwarded this newsletter?
LOVE IT.
If you liked what you read, consider signing up for your own subscription here:
