Omic.ly Weekly 57
January 6, 2025
Hey There!
Happy New Year!
And thanks for spending part of your week with Omic.ly!
This Week's Headlines
1) Radiation's got nothing on these bacteria!
2) lncRNAs and their role in epigenetics
3) How DNA codes for amino acids wasn't cracked until 1966
Here's what you missed in this week's Premium Edition:
HOT TAKE: With H5N1 Bird Flu on the rise, should we be preparing for pandemic 2.0?
Or if you already have a premium sub:
Extremophiles are giving us new tools to help combat contamination of pharmaceuticals.
They’re also helping us learn more about how to prevent oxidative stress!
Extremophiles are organisms that can live in what we consider extreme environmental conditions.
So, environments that are mostly inhospitable to life in general, such as places that experience extreme heat, extreme cold, drought or even radiation exposure.
The enzyme that powers the polymerase chain reaction was famously found inside of an extremophile bacteria that lived inside of the hot springs in Yellowstone National Park!
But the bug we’re going to talk about today, Deinococcus radiodurans, was originally discovered in 1956 due to its ability to withstand extreme radiation exposure.
It was found by Arthur Anderson during experiments where he was trying to figure out if gamma radiation could be used to prevent food from spoiling.
He exposed a tin of meat to levels of radiation known to kill all living organisms, and the meat still spoiled!
D. radiodurans was isolated soon after and has been studied ever since.
However, there’s been quite a bit of debate about how D. radiorans is able to survive doses of radiation up to 50,000 grays (Gy) - (3-6 Gy is fatal to humans!)
When organisms are exposed to Ionizing Radiation (IR), it causes double stranded DNA breaks and creates a ton of reactive oxygen species (ROS).
DNA breaks are bad news, but ROS can cause significant damage to DNA repair proteins, making them non-functional.
Luckily, D. radiodurans is very good at protecting the fidelity of those proteins and it had been hypothesized that this was because it also has the ability to soak up ROS better than many other bacteria.
It seems to do this using Manganese (Mg) as an antioxidant, but that’s not the full story, because Mg alone isn’t good enough to protect bacteria from extreme exposures to radiation.
But it was shown that E. coli (highly sensitive to radiation) could be protected from moderate radiation exposure if they were placed into cellular extracts from D. radiodurans.
An extensive analysis of those “Conan the Bacterium” cellular extracts showed that D. radiodurans produce a thick cocktail of peptides that appear to enhance the antioxidant effects of Mg.
Today’s paper explores how a synthetic peptide, DP1, (based on those found in D. radiodurans extracts) is able to protect other proteins from IR and the ROS that is created during that exposure.
It had been previously shown that DP1, in the presence of Mg and orthophosphate (Pi), acts as a superoxide dismutase (SOD - sciency name for ROS killer).
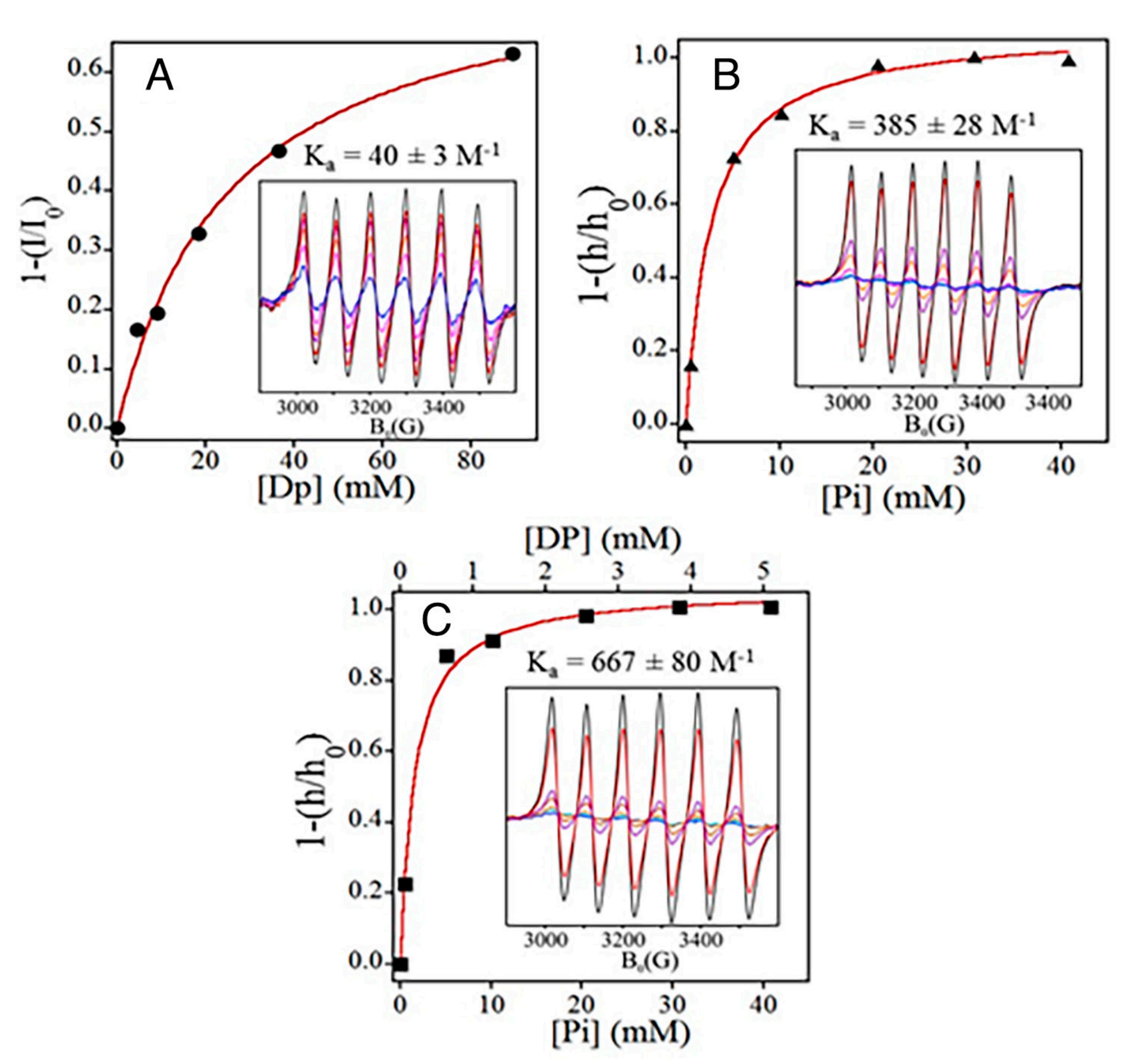
The figure above shows why this is the case.
The plots here display the Mg binding affinities of a) DP1 b) Pi and c) DP1 and Pi together.
DP1 weakly binds Mg (Ka=40), Pi binds it a bit more strongly (Ka=385) and DP1 and Pi when complexed together bind Mg very strongly (Ka=667).
This explains why DP1 and Pi act as such a strong antioxidant because they can trap a ton of Mg where it’s needed most!
DP1 based antioxidants are able to protect proteins from >60,000 gray of radiation exposure.
This is important because it means that extreme levels of IR can be used to decontaminate pharmaceuticals, or even be used to make killed whole cell vaccines that preserve the antigenic parts of bacteria and viruses while at the same time obliterating their nucleic acids with double stranded breaks!
###
Yang H, et al. 2024. The ternary complex of Mn2+, synthetic decapeptide DP1 (DEHGTAVMLK), and orthophosphate is a superb antioxidant. PNAS. DOI: 10.1073/pnas.2417389121
Long non-coding RNAs (lncRNAs): Epigenetics' latest layer of complexity!

Oh, epigenetics, why can't you just be simple?
Even your definition is complicated which is:
All of the non-sequence modifications to DNA that affect gene expression.
These include both DNA methylation and modifications to the histone proteins that coil up DNA into the chromatin that makes up our chromosomes.
These modifications alter the ‘state’ of chromatin which can be open and accessible for gene expression (euchromatin) or closed and inaccessible (heterochromatin).
The regulation of these two states is VERY important for determining what genes are expressed in our cells.
So, it stands to reason that these epigenetic modifications are super important!
Enzymes like DNA methylases are responsible for maintaining methylation, and a whole host of other enzymes regulate histone methylation, acetylation, phosphorylation, and sumoylation.
Together these are referred to as ‘marks’ and they determine whether stretches of chromatin are open or closed for gene expression.
These marks can also be dynamic, and often factors that are involved in regulating the establishment of these marks are implicated in human disease and cancer.
More recently, it has been recognized that another key player is involved in establishing and maintaining both open and closed chromatin:
RNA
While only 2% of the genome codes for proteins, about 75% of it is transcribed into RNA.
So, what's all of that extra RNA doing if it's not coding for protein?
Well, at least some of the time, it seems like these RNAs are helping to maintain the epigenome!
These non-coding RNAs fall into a couple classes, but the ones we're covering here are long non-coding RNAs or lncRNAs.
And the only criteria for getting into the lncRNA club is that you have to be >200 nucleotides and not code for anything!
But that doesn't mean these things aren't important.
Because we've discovered that lncRNAs play an essential role in bringing DNA methylases to locations in the genome where chromatin is tightly packed.
There is some evidence that they also attract histone modifying enzymes but their best known role is in regulating DNA methylation and establishing heterochromatin (closed/inactive).
One of the most studied lncRNAs is called XIST.
And it's responsible for fully inactivating one of the two X chromosomes in biologically female cells.
It does this by attracting methylases to one of the X's and this results in its systematic heterochromatization.
The ultimate goal of X-inactivation is to maintain dosage, or prevent female cells from creating too much of certain proteins!
lncRNAs operate similarly in other regions of the genome where they function to regulate gene expression.
While this isn't the only role that lncRNAs play in our cells, their epigenetic activities are important for making sure that the right proteins are expressed at the right time and in the right cells!
The structure of the DNA double-helix was published in 1953, but it took another 13 years to actually crack the genetic code.
The two key questions after the structure of DNA was figured out were:
1) How is it copied?
2) How does it code for proteins?
The first question was answered by Meselson and Stahl in 1958. DNA is copied ‘semi-conservatively’ - the old strands serve as a template for creating the new complementary strands.
The second question wasn’t fully resolved until 1966.
There, of course, were theories about how this occurred and Francis Crick gave a seminal lecture in 1957 (published as a paper in 1958), ‘On Protein Synthesis,’ which laid out the basic formula for how he believed this all worked.
He got it mostly right.
He proposed that there must be an 'adaptor' which carries the amino acids (we now call this transfer RNA, tRNA) to the template RNA (this became messenger RNA, mRNA) to synthesize the protein chain.
But Crick also hypothesized in this lecture that the genetic code is read 3 nucleotides at a time.
It was known that there were 20 amino acids and so Crick reasoned that since there are only 4 nucleotides in DNA and RNA, the ‘code’ for each amino acid couldn’t be 2 nucleotides (4x4) because that’s only 16 combinations, so optimally, it was a 3 nucleotide code (4x4x4) because that combination gave 64 possibilities - ~3 combinations for each of the 20 amino acids.
The first indication that Crick’s hypothesis was true came in 1961 when Nirenberg and Matthaei showed that an RNA sequence containing only U’s coded for poly-phenylalanine.
Inspired by this result, Crick showed that insertion or deletion of 1 or 2 bases causes proteins to become non-functional, but 3 base mutants still produce functional protein.
The code must be a triplet!
Nirenberg and Leder followed up in 1964 with triplet codes for a handful of amino acids but they frustratingly couldn't determine all of them.
That problem was solved by Har Gobind Khorana, an Indian American scientist.
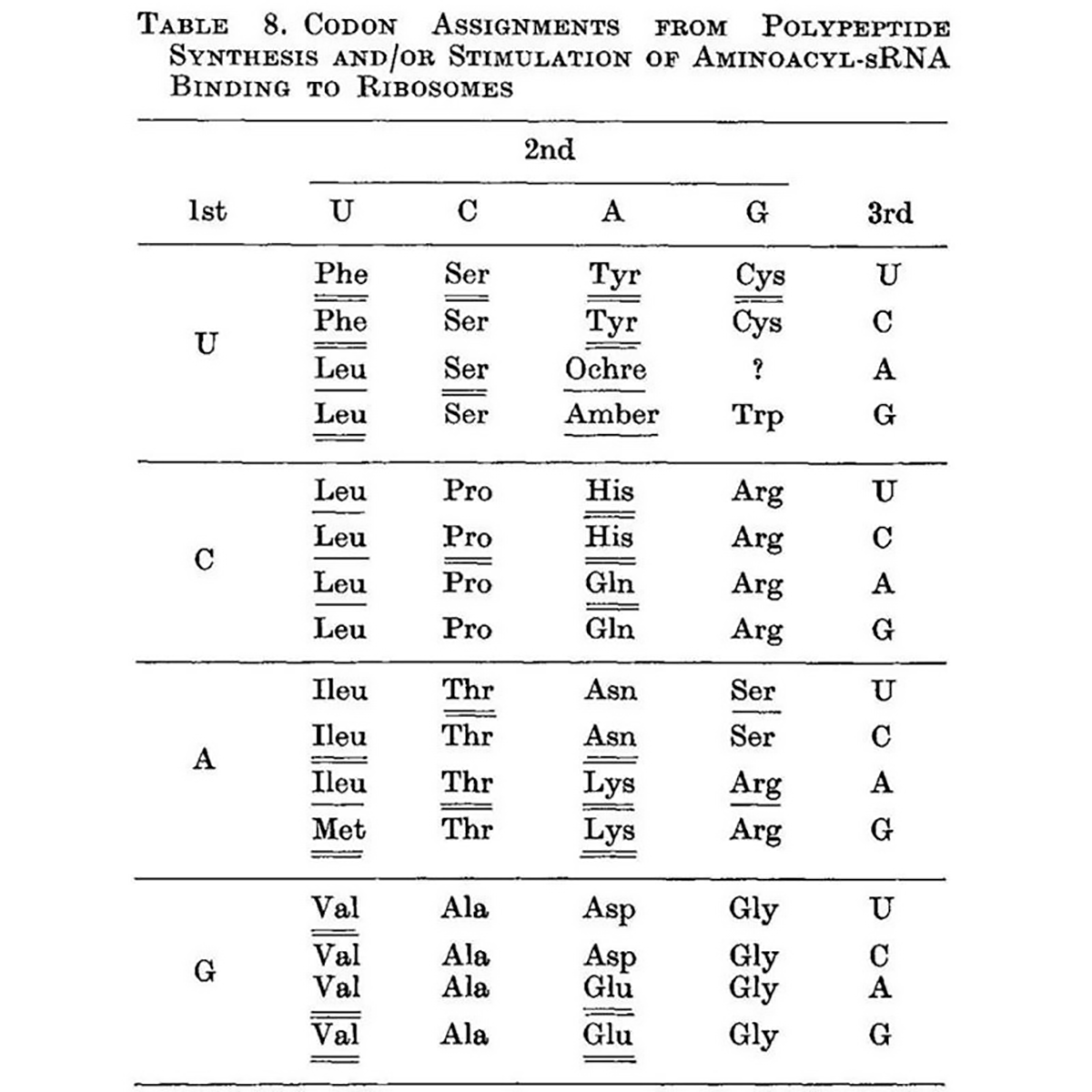
While Nirenberg and team were using binding assays to try to capture the RNA-amino acid associations, Khorana approached it differently by synthesizing repeating dinucleotide and trinucleotide RNA polymers and seeing which amino acids ended up linked together in the resulting protein chain.
The outcome of this work can be seen above (underlines), and this is the first presentation of a nearly complete codon table. The only missing assignment is UGA, the third and final stop codon.
Unsurprisingly, Khorana, Nirenberg and Holley (he determined the structure of tRNA) shared the Nobel Prize in 1968 for deciphering how nucleic acids code for proteins.
###
Khorana HG, et al. 1966. Polynucleotide Synthesis and the Genetic Code. CSHL Symposia on Quantitative Biology. DOI:10.1101/sqb.1966․031.01․010
Were you forwarded this newsletter?
LOVE IT.
If you liked what you read, consider signing up for your own subscription here:
