Omic.ly Weekly 56
December 30, 2024
Hey There!
Thanks for spending part of your holiday with Omic.ly!
This Week's Headlines
1) Science has figured out why Garfield is orange
2) Epigenetics is mostly about access
3) The most basic rule of DNA was first described in 1950
Here's what you missed in this week's Premium Edition:
HOT TAKE: Three omics predictions for 2025, and revisiting my predictions for 2024
Or if you already have a premium sub:
Thanks to some genetic sleuthing, we now understand why orange cats are orange.
Up until now, we only understood why 80% of orange cats were male!
And that’s because orange coat color in cats (and hamsters) is sex-linked.
Sex-linked traits are interesting because they don’t behave entirely the same way that traits that are linked to other chromosomes do.
For the regular old autosomes (the chromosomes who each have two identical copies), the genes that control traits come in pairs.
But with sex chromosomes, things get weird since the X chromosome and Y chromosome are different sizes and carry very different genes!
Biologically female mammals have two X chromosomes whereas biologically male mammals only have one.
This means that males who inherit damaged copies of genes from their moms almost always show a trait because they only get one copy!
But for females to show that same trait, they usually have to inherit two copies.
This simplistic logic holds true in the case of orange coat color in cats which has been shown previously to be linked to the X-chromosome, and this explains why 80% of orange cats are male!
However, we have had no idea how orange coat color in domestic cats was determined on the molecular level.
That changed recently when researchers discovered that orange coat color is caused by a deletion on the X-chromosome that alters the expression of a gene that regulates the production of the melanic pigments, eumelanin and pheomelanin.
Eumelanin is a black to brown pigmentation while pheomelanin is a reddish yellow color.
And it’s the production of these different pigments that give cats their colors and their interesting color patterns!
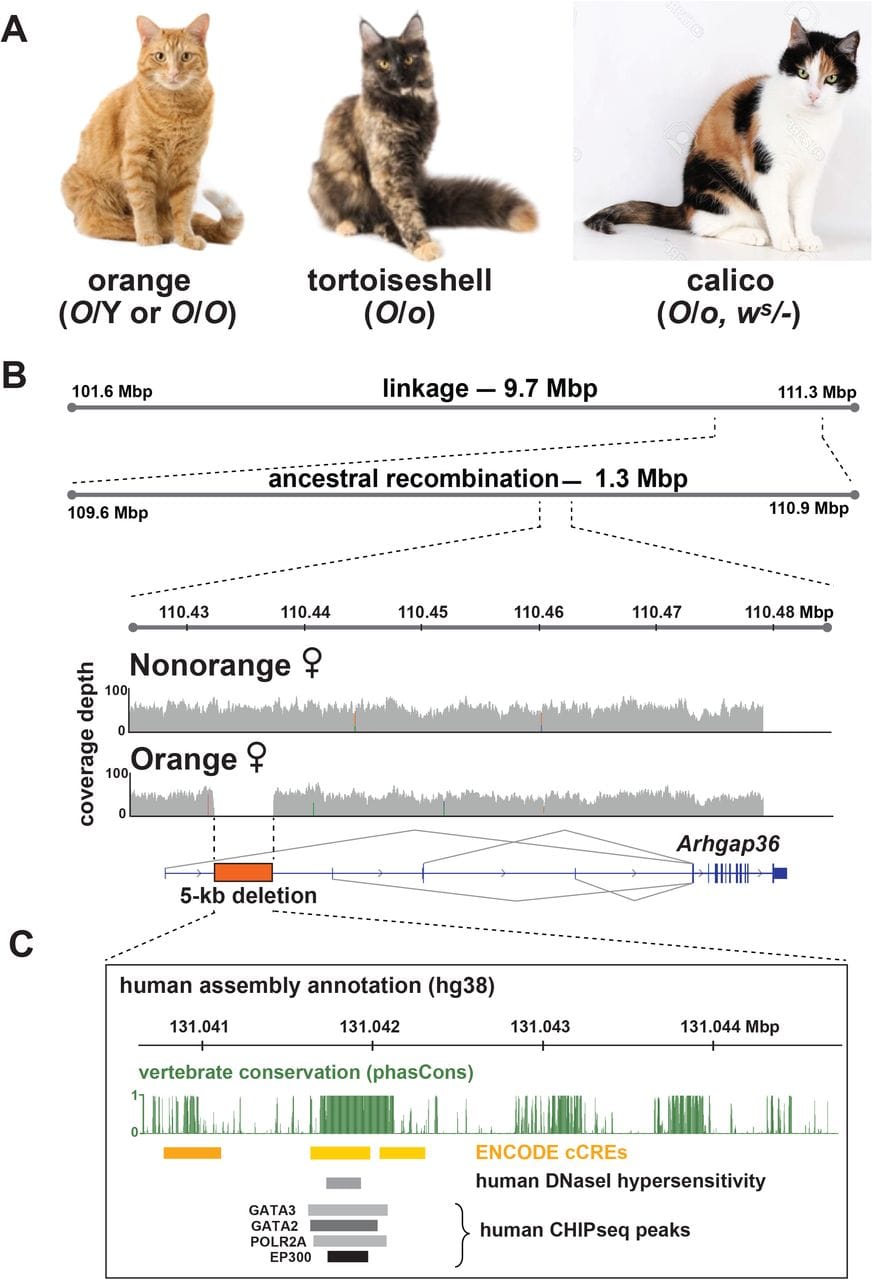
In the figure above the researchers show A) the different color patternings found in domestic cats which include sex-linked orange O/Y (male) and O/O (female), Tortoise shell and calico (which can be the result of X-inactivation based mosaicism and regular old autosomal inheritance) B) Genetic linkage mapping had previously shown that the orange trait was linked to a 9.7 megabase region of the X chromosome but more modern genetic techniques refined that to a 1.3MBp region and this paper further identified an 5kb deletion that was causal of orange coat color in felines. While this deletion doesn’t affect the coding region of a gene, it does occur in the non-coding regulatory region of one (see conservation map).
They went on to show that this gene, Rho GTPase Activating Protein 36 (Arhgap36), is upregulated 13 fold in the skin of orange cats.
They also showed that the activities of Arhgap36 result in the inhibition of protein kinase A (PKA), a gene essential for controlling the production of the melanic pigmentation proteins.
In the case of orange cats, this inhibition of PKA by Arhgap36 results in the underproduction of eumelanin (black) and the over production of pheomelanin (red/yellow == orange!).
The results of studies like this are important in the field of genetics because seeing physical traits is only half the story.
And, with respect to disease, if we don’t know the molecular mechanism behind the presentation of a trait, we’ll never be able to come up with treatments to deal with them in the clinic.
This is why basic science research (even in cats) is so important: it gives us the tools to help solve major molecular problems in humans.
Plus, knowing why Garfield is orange (molecularly) is just kinda cool!
###
Kaelin CB, et al. 2024. Molecular and genetic characterization of sex-linked orange coat color in the domestic cat. BioArxiv. DOI: 10.1101/2024.11.21.624608
Epigenetics and Genome Topology: What are DNA methylation and histone modifications actually doing?

Epigenetics is the study of all of the non-DNA sequence alterations that affect how genes are expressed within our cells.
These include methylation of cytosines in DNA and modifications to histone proteins.
Much of introductory biology exhibits DNA as a linear molecule, but in complex organisms, DNA is actually wrapped up around histone proteins to form a DNA-protein complex called chromatin.
And chromatin is what makes up our chromosomes!
Cytosine methylation and histone modifications work together to physically regulate access to the genetic code and control gene expression by condensing (closing) or relaxing (opening) chromatin!
The technical terms for these two states are heterochromatin (closed/inactive) and euchromatin (open/active).
This is further complicated by the fact that our chromosomes spend the majority of their time in the nucleus jumbled together like a giant ball of yarn.
But even this jumble of chromosomes is important!
Today we refer to this as ‘genome topology’ and the 3 dimensional structures that our chromatin forms in the nucleus may look like a totally random mess, but it’s actually ordered with the establishment of specific domains of activity!
These are of course regulated by epigenetic factors (proteins) that recognize and bind to epigenetic modifications on DNA and histones.
For example, the role of cytosine DNA methylation is to recruit histone methylases that then methylate histones and cause the formation of tightly packed heterochromatin.
This prevents gene expression from these regions.
But there are also a whole host of factors that are involved in regulating genome topology with four of the most important being CTCF, Cohesin, chromatin remodeling proteins, and Polycomb Group proteins.
These proteins are responsible for creating Topology Associated Domains (TADs) and help to define the boundaries between active and inactive chromatin!
CTCF (CCCTC-binding factor): Is a highly conserved protein that binds to DNA and marks TAD borders. This activity forms ‘loops’ in chromatin and in many cases facilitates long range interactions between enhancers and promoters.
Cohesin: Helps establish 3D chromatin architecture by working with CTCF to form loops. It is required for loop extrusion, and is important for maintaining chromatin organization.
Chromatin Remodelers: The most well known of these are ISWI and SWI/SNF and they push histones around to open or close access to the underlying DNA.
Polycomb Group (PcG) Proteins: Act as repressors of gene expression to keep chromatin closed and maintain cellular identities by preventing the inadvertent activation of genes that are not compatible with the function of a particular cell type.
But ultimately, the activities of these proteins are determined by the epigenetic status of DNA and histones!
A and T, and G and C are present in the same amounts in DNA. It’s the most basic rule of DNA. It’s also called Chargaff’s Rule.
Well before the structure of the DNA double helix was revealed by Watson and Crick (with the help of Franklin and Gosling!), biochemists first had to figure out the chemical make-up of deoxyribonucleic acid.
Identifying the bases and how they were linked together with a phosphate backbone was the work of Phoebus Levene.
He was the first to tease out the chemical makeup of ‘desoxypentose nucleic acid,’ which we lovingly refer to today as DNA!
However, because only 4 bases existed, Levene triumphantly declared that DNA was far too simple to be considered the chemical that stored the genetic information of organisms.
He fell into the same trap as most everyone else at the time, and believed that the genetic material was protein.
Levene proposed the ‘tetranucleotide hypothesis’ which stated that DNA was made up of repeats of the same 4 nucleotides and these nucleotides were present in equal amounts.
We now know that this was totally wrong and part of the reason why Levene thought that nucleotides were present in equal amounts was because the analytical methods used in 1928 were not accurate enough.
It took another 22 years, and the invention of much more sensitive paper based chromatography techniques to tease out the true chemical nature of DNA.
But the first cracks in the hypothesis that ‘proteins are the genetic material’ appeared in 1944 when Avery, Macleod and McCarty showed in a series of experiments that DNA, and not protein, could ‘transform’ the bacteria pneumococcus.
While the majority of scientists at the time ignored this paper (because the results were contrary to popular belief) Erwin Chargaff of Columbia University was intrigued.
He saw Avery’s results as the first evidence that maybe the field had it all wrong about DNA, and that it wasn’t just a random jumble of nucleotides but something much more important.
So, he did what he did best, got in the lab, extracted a bunch of DNA from a bunch of different organisms and calculated in exacting detail how much of each base was present in every sample he tested.
A good number of these papers are in German, but the one that I’m pulling from today was published in Nature in 1950.
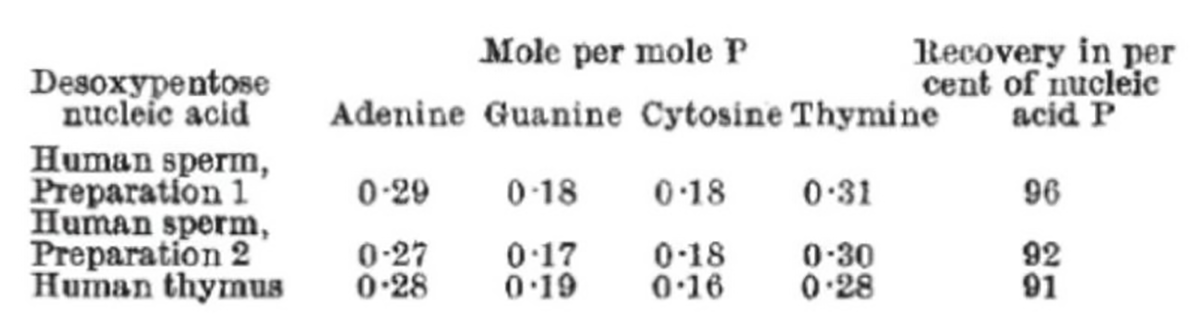
The figure above shows the nucleotide content of human sperm and human thymus.
~28% of human DNA is made up of Adenine, 18% Guanine, 18% Cytosine, and 30% Thymine.
These results show that the nucleotides aren’t all present in the same amounts as Levene had hypothesized, but that Adenine and Thymine, and Guanine and Cytosine are found in equal concentrations.
This was a key discovery on the path to showing how these seemingly complementary nucleotides bound together in the structure of DNA.
###
Chargaff E, et al. 1950. Composition of human desoxypentose nucleic acid. Nature. DOI:10.1038/165756b0
Were you forwarded this newsletter?
LOVE IT.
If you liked what you read, consider signing up for your own subscription here:
