Omic.ly Weekly 38
August 18, 2024
Hey There!
Thanks for spending part of your Sunday with Omic.ly!
I'll be on vacation from the 13th to the 25th. Unfortunately, edition 39 might be delayed until Tuesday the 27th, so don't be too worried if you don't get an email on the 25th.
This Week's Headlines
1) Need to reset your brain? It might be time to go on a trip
2) Being accurate about sequencing accuracy
3) Circulating cell free DNA testing was born in 1997
Here's what you missed in this week's Premium Edition:
HOT TAKE: Nabsys is back, baby!
Or if you already have a premium subscription:
Mr. Mackey always told us that, “Drugs are bad.” But sometimes they’re just misunderstood.
This is especially true when it comes to psychedelics and their therapeutic effects on the brain.
While many of these drugs have been vilified for their recreational use, there’s renewed interest in exploring the benefits of drugs like MDMA, LSD and psilocybin.
All of these drugs have been implicated in disrupting brain circuits which promote the feelings experienced during a psychedelic ‘trip.’
These can include visual alterations, changing colors or geometries, and even complex hallucinations of people, animals or environments.
What a user experiences during these trips can be highly dependent on their mood, where they are physically located, and their personality.
Users have described them as life altering and some emerge with a new sense of self.
And this is why psychedelics have been explored for therapeutic uses as a way to treat things like addiction, depression and post traumatic stress disorder.
But what happens on the molecular level during these events and how those changes affect the connections between neurons is largely unknown.
Psychedelics have been shown to activate the serotonin 2A (5-HT2A) receptor and alter how neurons communicate with one another by promoting the creation of new synapses in the medial frontal lobe and anterior hippocampus.
These changes in the hippocampus are thought to be what has the greatest therapeutic effect in depression because this is the region that influences a person’s sense of self.
Unfortunately, most of this work has been done in mice, and the specific effects of these drugs on human brains has not been studied in detail.
The authors of today’s paper were looking to change that and used functional magnetic resonance imaging (fMRI) to scan the brains of 7 participants who were given high doses of psilocybin.
Psilocybin caused widespread changes in brain connectivity (FC) across the cerebral cortex (the outer layer) and significantly increased desynchronization of brain activity.
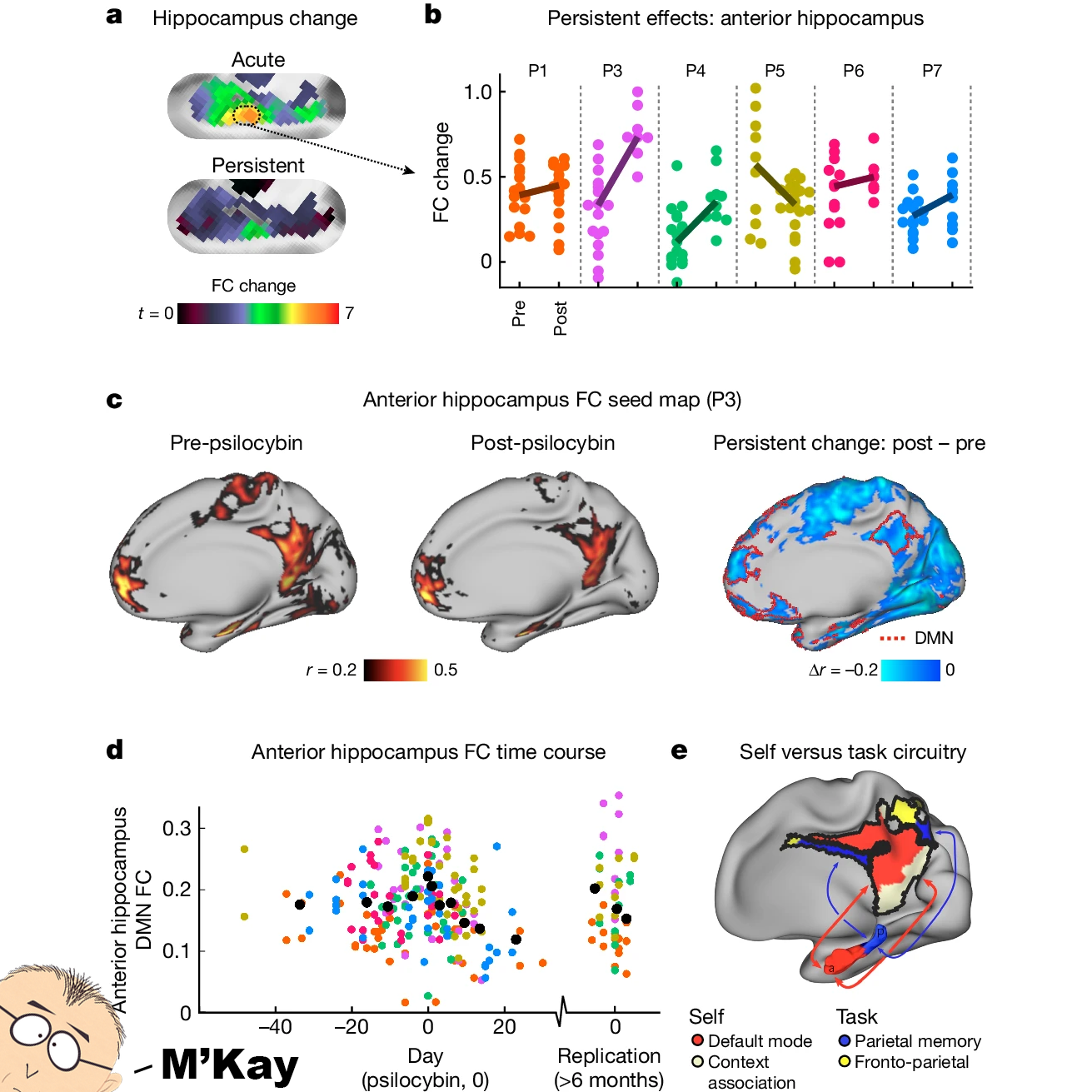
But the most interesting result can be seen in the figure above where a persistent effect was observed in the anterior hippocampus a) at psilocybin dosing (acute) and 3 weeks later (persistent). b) Changes were seen in all 7 participants. c,d) There was reduced connectivity between the hippocampus and the default mode network (DMN - regions involved subconscious activities and strongly linked to depression) e) A model of hippocampal and DMN circuits.
Or, TL; DR...psilocybin is able to disrupt key brain circuits involved in depression and this explains why it and other psychedelics promote lasting effects in depressed brains.
This could lead to tailored therapeutic programs that prescribe psychedelics based on brain activity pre- and post- treatment.
Maybe drugs aren't so bad.
###
Siegel JS, et al. 2024. Psilocybin desynchronizes the human brain. Nature. DOI: 10.1038/s41586-024-07624-5
High throughput sequencing metrics: Let’s be accurate about accuracy.

If “What is a genome?” is the most loaded question in the world of short-reads, the most loaded question for long-reads is “What’s the accuracy?”
A lot of this comes down to the semantics that PacBio and Oxford Nanopore (ONT) use when talking about accuracy.
Because in conversations they tend to use the different types of accuracy essentially interchangeably.
There are 3 main types of accuracy in sequencing:
Base Accuracy - The accuracy of each individual base call. This is the traditional Phred Q-score and a general measure of quality used for assessing most short-read sequencing outputs. Element is Q40+ (99.99%), Illumina/Complete is Q30+ (99.9%), and Thermo is Q20+ (99%).
Read Accuracy - Up one level from base accuracy, what percentage of the bases in a read are accurate. PacBio HiFi reads are 99.9-99.999% accurate, ONT is 70-99.9% accurate.
Consensus Accuracy - The accuracy of the consensus sequence, or, the accuracy after assembling all of the data using multiple reads to correct all the errors.
Before PacBio released HiFi reads, both PacBio and ONT required 'polishing' to produce useful datasets. Reads where ~25% of the bases are wrong are challenging to use for most sequencing applications.
Polishing requires the use of short reads to correct all of the errors because of their very high accuracy, and so long-reads struggled to gain traction beyond niche genome scaffolding applications since these hybrid sequencing approaches are very expensive.
Another expensive method for correcting these sorts of errors is to just do more sequencing.
This works for PacBio because the errors in their data are random. This is why HiFi circular consensus sequencing (CCS) has such high read accuracy, it’s a self-polishing method!
It also works for some ONT data errors, but their method is prone to systematic errors (always happen in the same sequences), so sequencing more will not remove those and requires short-read polishing methods to be fully resolved.
More recently, ONT have touted read accuracies at the same level as PacBio, with sequences for HG002 coming off their dual reader R10 pores with a 'duplex' read accuracy of 99.9%+.
But, at the end of the day, the key thing to know about accuracy is that traditional base accuracy is more relevant for short-reads when addressing quality and read accuracy and consensus accuracy are more important for long-reads.
This will become more apparent as each of the two technologies settle on the niches where they excel - long-reads for high quality genomes, and short-reads for just about everything else!
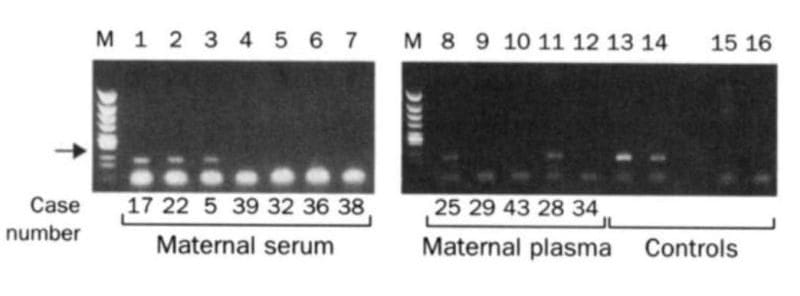
The two gels below spawned a multi-billion dollar industry that didn't exist prior to their publication in 1997.
While it may seem obvious today, you might find it surprising that we didn't know that fetal DNA was present in a pregnant mother's bloodstream until the late 1990's.
Prior to this discovery, genetic testing on fetuses was only performed if a problem was suspected with a pregnancy or if there was a family history of genetic disorders.
This testing was performed by karyotyping fetal cells.
You've probably seen one of these 'chromosome spreads' in a biology textbook where each of the 23 pairs of chromosomes are lined up next to one another.
This allows a geneticist to check that the chromosomes are intact and see if there are any abnormalities such as those found in Down Syndrome where patients have 3 copies of chromosome 21 instead of 2.
But, back in 1997, the process for collecting fetal cells was very invasive and was done using one of two techniques:
Amniocentesis (Amnio) - a 3-5" long needle is inserted into the mother's abdomen to collect 20 milliliters of amniotic fluid for testing.
Chorionic Villus Sampling (CVS) - A 6" long needle is guided via ultrasound, either vaginally or through the abdomen, to obtain tissue from the placenta for testing.
Unfortunately, these procedures carry a risk and 1-2% of the time they can lead to the loss of the fetus.
Recognizing that there had to be a better way, Yuk-Ming Dennis Lo set about figuring out how to get at fetal DNA without having to perform Amnio or CVS.
He knew that fetal cells made it into the maternal bloodstream and that mother and child exchanged cellular material.
But he couldn't isolate enough fetal cells from the blood to do prenatal genetics with them.
Luckily, in 1996, Lo heard that a team in Switzerland had shown that tumors actually shed cell free DNA into the bloodstream and this tumor DNA could be detected using PCR and primers specific to the tumor DNA.
He reasoned that a baby, or more accurately, the placenta, was basically like a giant tumor and shared the bloodstream with the mother.
So, it made sense that it too should shed cell free DNA into the bloodstream.
The figure above is proof that Lo's hypothesis was correct and fetal DNA could be isolated and amplified from the blood of pregnant females.
In this figure, the arrow highlights a 198bp PCR product from the Y chromosome. Case numbers greater than 30 are pregnancies with female fetuses (don't have a Y), and case numbers less than 30 are male fetuses (have a Y).
Further technical advancements and the invention of high throughput sequencers have transformed this discovery into a popular pregnancy screening test.
Today, non-invasive prenatal testing is second only to oncology testing in sequencing market share.
###
Lo YMD, et al. 1997. Presence of fetal DNA in maternal plasma and serum. The Lancet. DOI:10.1016/s0140-6736(97)02174-0
Were you forwarded this newsletter?
LOVE IT.
If you liked what you read, consider signing up for your own subscription here:
