Omic.ly Weekly 36
August 4, 2024
Hey There!
Thanks for spending part of your Sunday with Omic.ly!
This Week's Headlines
1) If only Ponce de Leon had known about IL-11
2) Why short-read sequencing can't ever get us a 'whole genome'
3) Frederick Sanger sequenced the first DNA genome, he didn't use 'Sanger Sequencing' to do it
Here's what you missed in this week's Premium Edition:
HOT TAKE: After an IPO valuing them at $3.5B 3 years ago, 23andMe wants to go private for $200m, signaling another tremendous SPAC failure
Or if you already have a premium subscription:
Who knew that the fountain of youth was as simple as blocking inflammatory signaling through IL-11?
At least that’s what a recent paper has discovered in mice.
Humans have been trying to find themselves cures for what ails them since the beginning of time.
And it’s basically why the entire healthcare industry exists.
We’re all looking for ways to lead happier, healthier and longer lives.
This began in olden times with ‘healers’ mixing up potions that were purported to cure disease (or expel demons).
We still have those types of people.
But we also have science based approaches now that allow us to see if any of these treatments actually work.
While health and aging research has existed for as long as ‘healers’ have, we’ve seen an explosion of scientific research in this field in the last few decades.
We’re now starting to hone in on the genes and pathways that can be exploited to improve our health and lifespan.
Some of this comes down to us doing the things that we know we should be doing, like not eating too much junk food, getting exercise, being social and spending more time outdoors.
But there are also very likely to be things we can manipulate on the molecular level to extend the benefits we get from these activities even further!
And studies looking at changes that occur in our bodies as we get older have identified increased inflammation as a hallmark of aging.
Research in worms, flies and mice have shown that lifespan can be increased by inhibiting some of these pathways.
One of the best studied of these is the mTOR (mammalian target of rapamycin) pathway and mTOR is a protein kinase (phosphorylates things, usually to turn them on) involved in regulating gene expression, cell growth and metabolism.
It can be activated through a number of pathways including by pro-inflammatory cytokines like IL-11.
The authors of today’s paper wanted to see what would happen to mice in the absence of IL-11 signaling and hypothesized that since IL-11 increases with age, its inhibition might be able to reverse some effects of aging.
And that’s exactly what they saw!
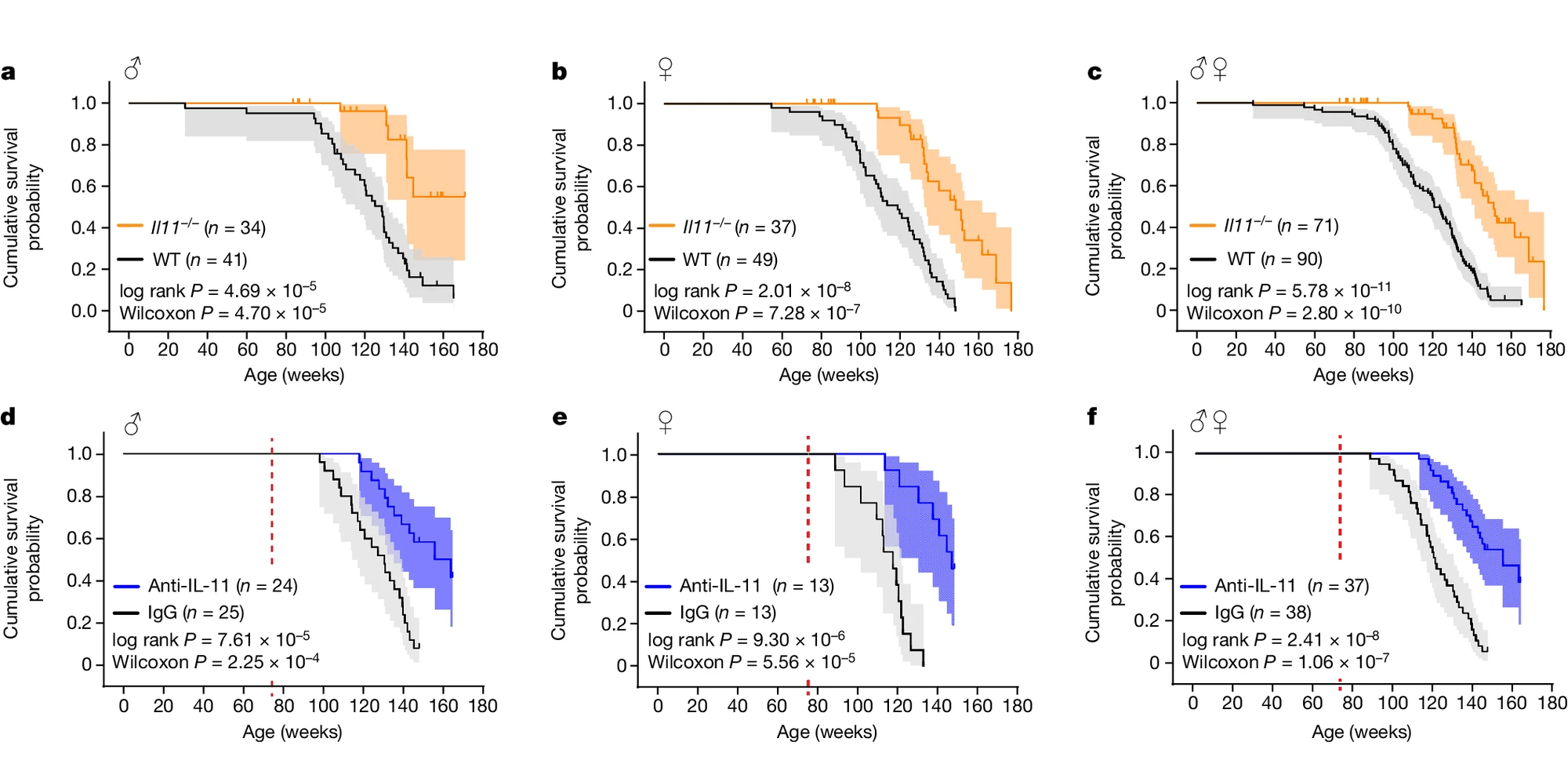
In the figure a, male (a) and female (b) mice who had the IL-11 gene deleted lived, on average, 20% longer than their non-mutant counterparts and this effect was mimicked by an antibody that blocks IL-11 (d,e,f).
But unlike other drugs in this space (like rapamycin) that only improve how long the test subjects live (but also with some pretty nasty side effects), this therapy appeared to make the mice healthier without any observed problems.
It’s important to keep in mind that this is early research in mice, but the authors point out that IL-11 therapies are being tested in the clinic for other disease states.
So, we could have some initial data on the health and lifespan effects of IL-11 inhibition in humans relatively quickly.
###
Widjaja AA, et al. 2024. Inhibition of IL-11 signaling extends mammalian healthspan and lifespan. Nature. DOI: 10.1038/s41586-024-07701-9
Short-reads miss things, or: How I learned to stop worrying and love the long-read.

Chromosome 21 is 47 million contiguous bases and Chromosome 1 is 247 million contiguous bases - the other 22 fall somewhere in between.
When we sequence a genome using short-reads, the first step in the process after DNA isolation is Fragmentation.
We take those big, beautiful, contiguous bases and chop them up into 200-300 base pair fragments.
I wonder if we lose important information when we do that?
We do!
And this is because when we 'align' those hundreds of millions of 300 base pair fragments, we’re using the unique overlapping sequence within those fragments to figure out where they belong in the genome.
It’s like putting together a huge puzzle!
But what happens when those fragments aren't unique?
20% of the genome has repetitive sequence longer than 200bp, so we’re left with millions of fragments that can’t be aligned to the right places.
This becomes a big problem in disease diagnosis because if we don’t know the orientation or location of these fragments, or worse, they’re mapped to the wrong places, we can totally miss important indicators of genetic disease.
Short-reads are especially problematic for:
Pseudogenes - Viruses liked to infect our ancestors. Sometimes they copied and repeated adjacent sequences as they entered and exited our genome. Except these copies now exist in a genetic no-man’s land where they aren’t expressed, and they serve only to confuse and deceive.
Translocations and Inversions - These happen when genetic content is rearranged and put back in the wrong place (translocation) or flipped (inversion). These errors can have a huge impact on gene regulation, especially if unbalanced or fused, and we mostly miss them with short reads.
Repeat Expansions - Trinucleotide repeat expansions are causal of 50 genetic diseases including Huntington’s, Fragile X and spinocerebellar ataxia. ALS can also be caused by a repeat expansion. Many of these are too long to be resolved with short-reads and so archaic techniques like Southern blot are used to measure them.
Variant Haplotyping and Phasing - You have two of each chromosome, the variants between those copies are called alleles. Knowing which alleles the different variants are on is important for understanding a number of diseases, especially those caused by compound heterozygotes (two different mutations in the same gene spaced very far apart).
Mitochondrial Genome - Did you know your cells actually have two genomes? A lot of the content of your mitochondrial genome can be repeated in your nuclear genome. But the copies in your nuclei don’t matter.
How can we capture all of this information to make sure we’re getting an accurate representation of the whole genome without missing anything important?
We can use high quality long-reads, and luckily for us, we have a few good options to fill in these short-read gaps!
Frederick Sanger invented a famous DNA sequencing method. It's not the one pictured below (which he also invented).
Sanger is one of the few scientists to be awarded more than one Nobel Prize.
His first was for determining the amino acid sequence of Insulin!
His second was shared with Walter Gilbert and Paul Berg for developing nucleic acid sequencing methods.
To this day, 'Sanger' sequencing is a gold standard method in genetics.
And despite the advent of high throughput sequencing technology, millions of Sanger reactions are still performed every year!
But Sanger has another claim to fame.
He sequenced the first DNA genome which was the bacteriophage PhiX174.
This may sound familiar.
Because PhiX is used widely as a sequencing control!
This is partly to memorialize that it was the first, but mostly because it has a similar number of AT and GC bases.
But, Sanger didn't use his eponymous sequencing method to sequence it!
This came as a bit of a shock to me.
And my discovery of this fact went something like this:
“What the (expletive) is the ‘plus and minus’ method!?”
And so, I'm not going to cover Sanger's sequencing of bacteriophage PhiX174.
Instead, we'll review the method he used to do it!
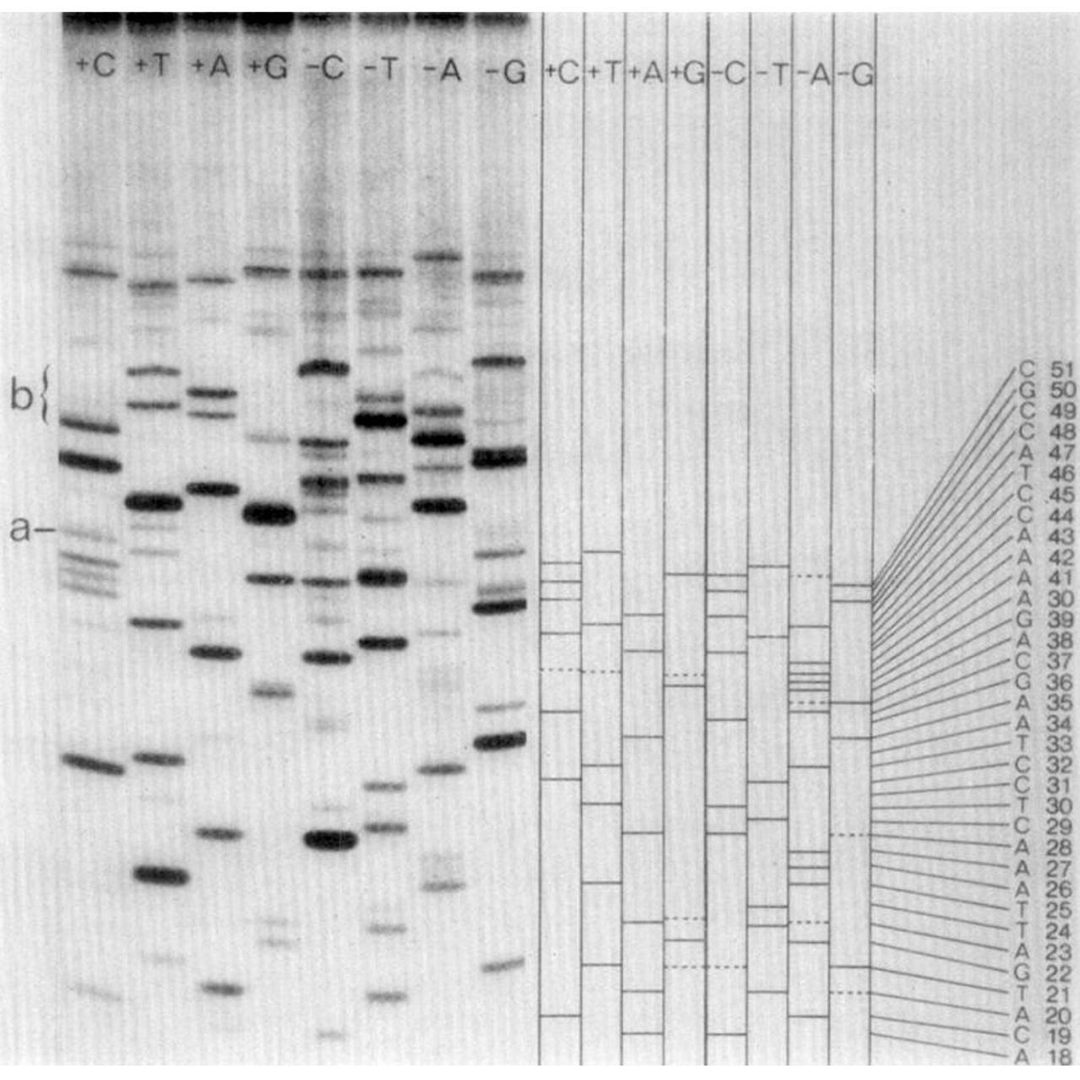
So, what exactly is the 'plus and minus' method?
It can be seen above.
But don't be fooled!
While it looks like regular old gel based Sanger sequencing, there are actually twice as many lanes!
And they're labeled ‘+C+T+A+G’ and ‘-C-T-A-G.’
Are you starting to see where the 'plus and minus' name came from?
The minus lanes are made by priming the thing you want to sequence with a restriction fragment, adding all the bases and a polymerase, and then stopping the reaction.
Those random products are then purified and added to a new reaction that contains only 3 of the 4 bases.
Whatever additional extension occurs stops at the position of the missing base.
So, the band in the gel is -1 from where the -base should be!
The plus lanes are made using a similar process:
Random fragments are generated and then the purified fragments are added to a new reaction containing only 1 of the nucleotides along with T4 polymerase.
T4 polymerase is special because it will 3’ degrade DNA, but interestingly, it will stop degrading DNA at “residues corresponding to the one [nucleotide] that is present.”
Translated: if you only add adenine nucleotides, it creates fragments that all end in adenine.
So, when you run these 8 reactions (+C+T+A+G -C-T-A-G) out on a gel, you can use the plus and minus signals from each base to determine the sequence of the fragment that was present!
Two years later, chain terminating dideoxynucleotides were added to this scheme, allowing 'Sanger' sequencing to be done in 4 lanes instead of 8 and with fewer steps and artifacts.
###
Sanger F, Coulson, AR. 1975. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. JMB. DOI: 10.1016/0022-2836(75)90213-2
Were you forwarded this newsletter?
LOVE IT.
If you liked what you read, consider signing up for your own subscription here:
