Omic.ly Weekly 33
July 14, 2024
Hey There!
Thanks for spending part of your Sunday with Omic.ly!
This Week's Headlines
1) Parkinson's Disease gets a new risk factor
2) Sequencing libraries and strategies for doing quality control
3) Defenders from the sea: Our bloody history with horseshoe crabs
Here's what you missed in this week's Premium Edition:
HOT TAKE: Element is off to the races with a new raise while Illumina plays catch-up on single-cell
Or if you're already a Premium Subscriber:
A recent survey of 2,000 families with Parkinson’s Disease has turned up a new risk factor: a variant in RAB32
Parkinson’s Disease (PD) is a neurodegenerative disease with the majority of cases occurring in people over the age of 60.
Early signs of the disease are tremors, difficulty to balance and slowness with dementia, mood swings and behavioral changes presenting at later stages.
85% of cases are sporadic, meaning there’s no family history of disease, and the absolute cause of Parkinson’s is still largely unknown but environmental exposures to certain pesticides, heavy metals, and genetic factors have all been linked to disease.
10-15% of cases are familial or inherited, but even in cases of familial PD it’s been very hard to track down a specific cause of the disease.
The molecular hallmark of PD is the development of Lewy bodies in neurons which contain aggregations (clumps) of the protein alpha-synuclein (SNCA) and this clumping is thought to lead to neuronal death.
To date, 90 risk variants have been identified along with a few notable genes like SNCA, LRRK2, and PRKN.
These genes play a variety of different roles in cells including functioning in cellular metabolism, cell death and protein decay indicating that the cause of PD can be multifaceted.
A subset of these mutations have the clinical presentation of PD, but at autopsy do not have SNCA aggregates in neurons.
This, of course, adds to the frustration of not knowing how best to treat the disease because most PD research has focused on how SNCA aggregation leads to cell death.
There appears to be much more to this story.
As for LRRK2 (Leucine-rich repeat kinase 2), it’s a small kinase that phosphorylates a gene called Parkin (PRKN) that’s involved in degrading proteins.
Increased activation of LRRK2 is associated with neuronal cell death in cellular and animal models of PD.
And while we don’t know exactly how or why this causes PD, the LRRK2/PRKN pathway appears to contribute to the disease in significant ways.
That’s why the recent discovery of a new gene involved in PD was so interesting, because it directly impacts the activity of LRRK2.
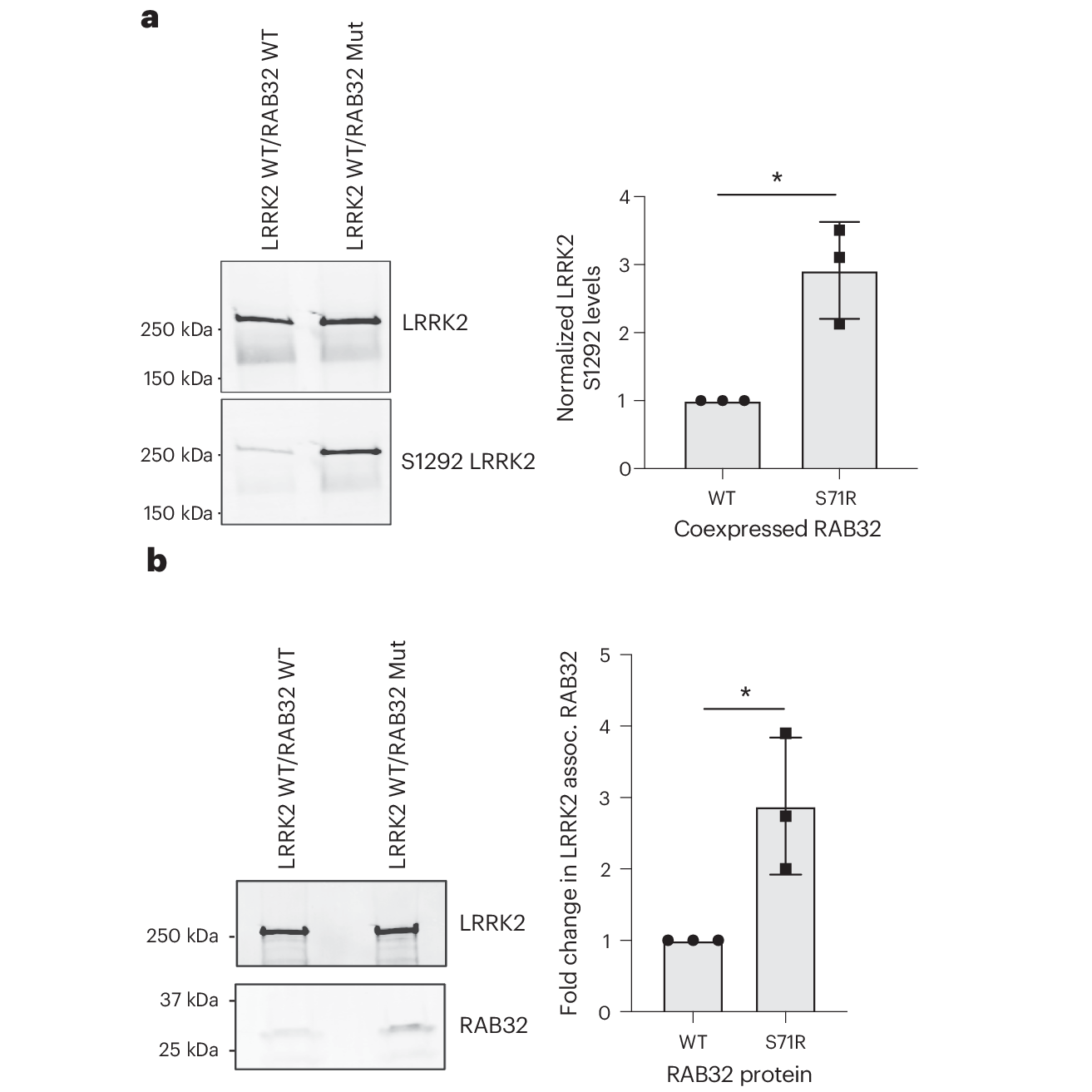
In this research, a family based case-control analysis of 2,184 PD families and 69,775 controls revealed that a variant of RAB32, a protein kinase, was significantly associated with PD (odds ratio 65.5!)
In the figure above you can see that this mutant a) results in a significant increase in LRRK2 S1292 phosphorylation and b) associates much more strongly with LRRK2 than the non-mutant.
This variant is causal of PD in only about 1% of familial cases, but discoveries like this help us to better understand the molecular mechanisms that lead to PD and offer potential targets for therapeutic development.
###
Hop PJ, et al. 2024. Systematic rare variant analyses identify RAB32 as a susceptibility gene for familial Parkinson’s disease. Nature Genetics. DOI: 10.1038/s41588-024-01787-7
You can't do sequencing without libraries. But you don't know if you have libraries unless you look at them.

There's nothing more nerve wracking than loading your samples on a sequencer and waiting to see how the metrics look.
Way back in olden times, there was a thing called the 'first base report,' and after screwing in your reagent bottles, pulling your flowcell off the cBot, and oiling up your prism, you got to wait a couple hours for a window to pop up telling you if anything you spent the last days of work doing actually worked.
Kids these days just fill up the drawers and walk away BUT one way to know if your sequencing run might actually work is to do quality control checks throughout the preparation process!
Some vendors will tell you they've invented 'easy buttons' and between normalizer beads and their 'simple to use kits' you can skip most of these steps and pray that you don't blow $1,000-25,000 finding out that Joey added another round of 80% EtOH instead of water after the last bead clean.
Trust me, you want to know that before you load your 'samples.'
The key quality control steps during sample preparation that I've always implemented in all of my processes are:
Post-Extraction Quantification (Quant) - You sequence nucleic acids and to get nucleic acids you have to purify them from a sample. The sample can be anything from saliva to semen, but you want to know that you got the good bits out of that process before moving on to turning it into a library. For this quant I suggest using picogreen because it's quick, easy and pretty darn accurate. At this step it serves two purposes: 1) it tells you if your extraction worked, 2) you can use that number to normalize the amount of sample that goes into library prep!
Post-Library Fragment Analysis - It's debatable whether this should be done EVERY time. I've made the argument that it should because it saves a good number of failed runs but it adds a couple bucks to every sample. This is usually done using a Bioanalzyer, Tapestation or AATI HPLC system but basically what this does is tell you the fragment sizes that are contained within your library. For short-read sequencing, you generally want these to be peaking around 400-500bp for 150x150 runs. It also shows you the fragment size distribution.
Post-Libray and/or Post-Capture Quant - The quant you do before you load your samples on the sequencer should be the most reliable and accurate quant you can use. Good options here are qPCR or ddPCR based quants that use the sequencing primers as the targets for quantification because this tells you how much sequenceable library is in the sample and not just how much total DNA is in there. Those are two different things!
But chances are you're going to have a few 'where the F*$# did my library go?' moments.
And it's best to know that before you try to sequence them.
Horseshoe crab blood is our first line of defense against endotoxin contamination of surgical tools and medical devices. Seriously.
We figured out pretty early on that sticking unsterilized medical equipment into people during surgeries usually ends badly.
And badly here means that a lot of people died from bacterial infections.
"Isn't that why we use an autoclave to sterilize those things?"
Yes, but that's only half of the solution.
Because gram negative bacteria also produce endotoxins like Lipopolysaccharide (LPS) which make up their cell wall.
Autoclave sterilization kills bacteria but has no effect on LPS which is a potent stimulator of the immune system and can lead to septic shock.
This can be caused by the Shwartzman Phenomenon which was first discovered by Gregory Shwartzman in 1926.
He found that injecting rabbits with two doses of gram negative bacteria (have an endotoxin laden cell wall) but not gram positive bacteria (no endotoxin) led to severe, hemorrhagic necrosis at the injection site - or, the immune system reacted, clotted the area, and caused the cells in that location to die.
You can see why endotoxin contamination of medical devices and other things that make contact with blood might be a bad thing.
So, for a long time, manufacturers of these things kept large colonies of rabbits and used them as endotoxin detectors.
At the end of manufacturing, devices would be washed with a saline solution and that would then be injected into a rabbit. If the rabbit got a fever, it meant the device was contaminated.
However, the rabbits got a reprieve in 1956 when Frederick Bang made a crazy discovery while studying the horseshoe crab Limulus polyphemus.
One of his crabs got sick, became lethargic and died. Upon closer inspection, he noticed that the crab's blood turned into a thick gel.
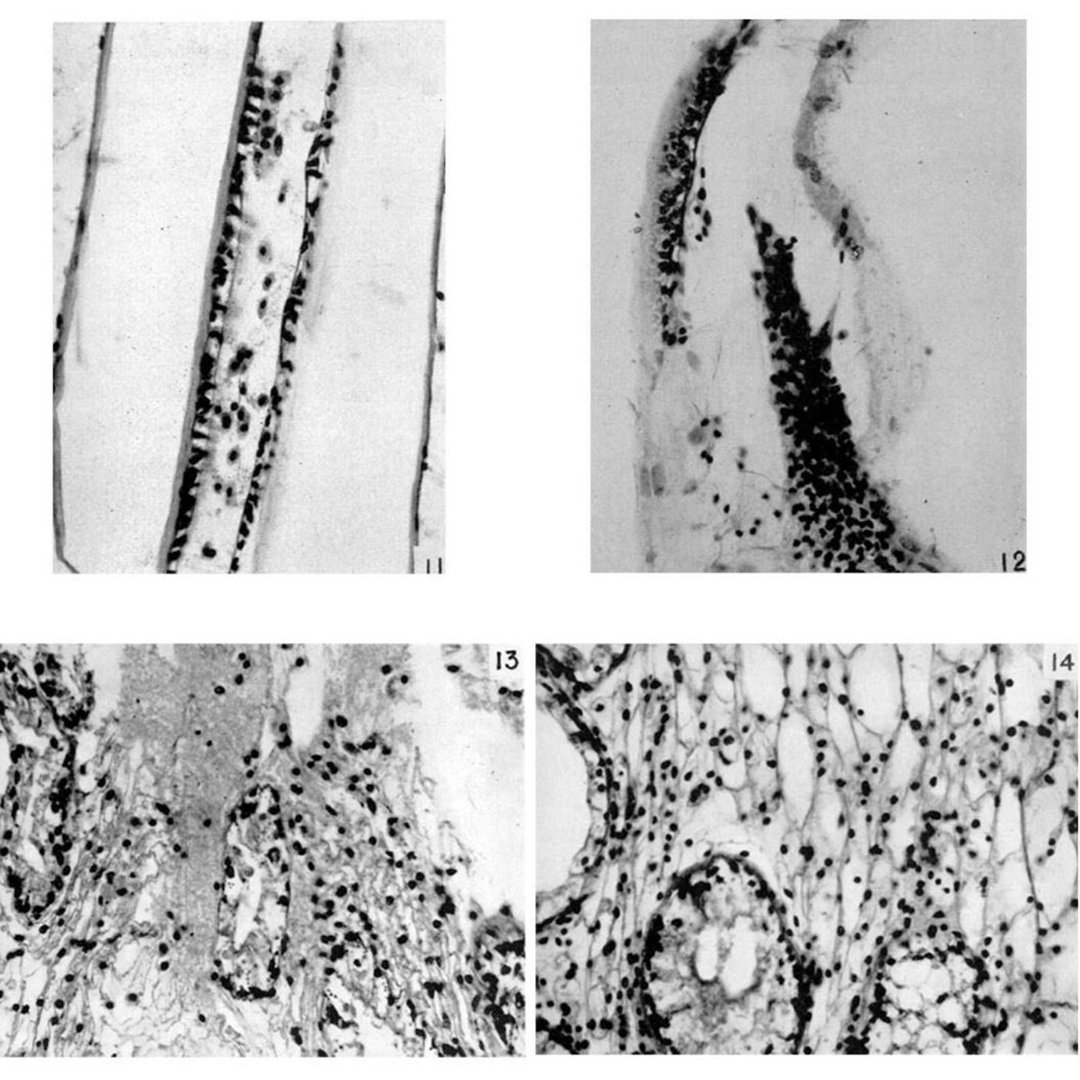
Doing what any curious scientist would, he cultured the blood, saw gram negative bacteria (endotoxins!), and injected the cultures into a bunch of other crabs.
The figure above is a compilation of the 'before and after' of those injections. Figures 11 & 13 are 'before,' and 12 & 14 are the clotted 'after.'
Bang, thinking this looked a lot like the Shwartzman Phenomenon on steroids, hired Jack Levin, a hematologist (blood nerd) to study this effect further.
What Bang and Levin discovered was that horseshoe crab blood cells, called amoebocysts, are highly sensitized to endotoxins and secrete a massive amount of clotting factor in an attempt to stop an infection!
It was quickly realized that a test based on horseshoe crab blood would be WAY cheaper than maintaining rabbit colonies and in the 1970's the FDA approved the Limulus Amoebocyte Lysate Assay for this use.
More recently, regulatory changes were proposed in 2023, enabling the use of a synthetic version of this test, and should reduce the bleeding burden of our crab friends.
###
Bang FB. 1956. A bacterial disease of Limulus polyphemus. Bull Johns Hopkins Hosp. PMID: 13316302
Were you forwarded this newsletter?
LOVE IT.
If you liked what you read, consider signing up for your own subscription here:
