Omic.ly Weekly 32
July 7, 2024
Hey There!
Thanks for spending part of your Sunday with Omic.ly!
If you're enjoying what you're reading, please feel free to share it with your friends and colleagues who might get a kick out of it too!
This Week's Headlines
1) Bridge RNAs are here to take CRISPR's crown
2) The messiest part of Omics? Dealing with all the different sample types
3) In 2009, high throughput sequencing gave us our first glimpse of the 3-D organization of the DNA in our nuclei
Here's what you missed in this week's Premium Edition:
HOT TAKE: One of the most underappreciated techniques in Omics is Hi-C. That needs to change
Or if you already have a Premium Subscription:
Move over CRISPR, bridge RNAs are here to make the precision genome edits that you can't.
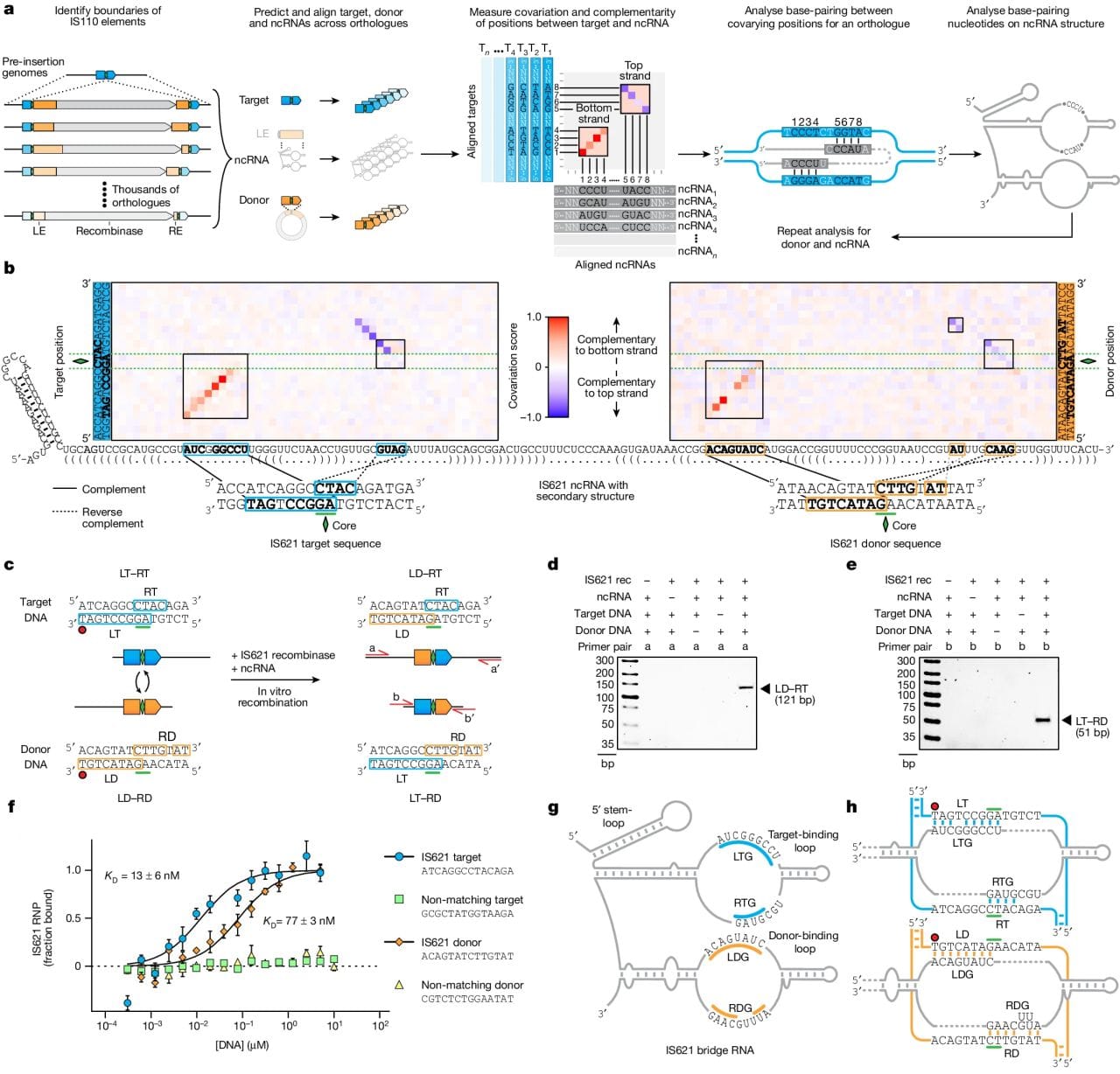
A lot of noise has been made about genome engineering in the last two decades.
But the idea of engineering genomes really began in the 1980’s with the discovery of mammalian, viral, and bacterial recombination systems.
In the early days we envisioned evolving these tools to precisely manipulate large pieces of DNA within living cells.
We quickly realized that was very hard and were only really able to do it reliably in embryos or in plant seeds.
Large scale engineering in living organisms would have to wait.
But then along came RNA interference, TALENs and their friend CRISPR which allowed us to do some of those cool things we had initially imagined, just at a smaller scale.
RNAi allowed us to chop up RNA’s we didn’t want around and prevent the expression of specific genes.
TALENs and CRISPR allowed us to make single nucleotide edits or create small insertions and deletions in specific genes.
Some of us even attempted to use CRISPR to insert new sequences into specific places in the genome.
But that process is very messy, highly inefficient and really only works in situations where you can take cells out of someone’s body, edit them, make sure those edits are correct, and then put them back in.
Wouldn’t it be nice if you could just take a big piece of DNA that you want in a new place and use an enzyme to efficiently stick it in there without all of this extra fuss?
We might be in luck.
Because a new genome editing system has been identified in bacteria that uses a small guide RNA and a recombinase to specifically insert a donor DNA sequence into a target.
And the entire system is available in a tiny package that should be slightly easier to get into cells than the other competing alternatives.
The figure above shows how it was discovered and how it works. a) bioinformatic identification of guide RNA sequences found in IS110 recombinase family members. b) heat map identifying two sets of highly conserved regions. c,d,e) this system is able to perform a recombination reaction in a test tube. f) In the presence of its guide RNA, IS621 (an IS110 family member) has very high affinity for target and donor sequences. g) a graphical representation of how IS621 uses its guide RNA to ‘bridge’ the donor and target DNA together.
The authors went on to show that this system is programmable and can make specific edits.
And, interestingly, because the IS110 family is so large, there are a number of recombinase/guide RNA combinations to explore that might provide additional functionality or increased edit specificity.
Bridge RNAs look like a promising new tool in our genome engineering arsenal that might someday allow us to make the edits we've always dreamed of.
###
Durrant MG, et al. 2024. Bridge RNAs direct programmable recombination of target and donor DNA. Nature. DOI: 10.1038/s41586-024-07552-4
The good, the bad, the...uhhh: Everything you ever wanted to know about all the different Omics sample types!

But first, what's a 'sample type?'
It just refers to the source material being tested!
For molecular testing, this is usually a body fluid or something that came in contact with a body fluid!
Blood tubes:
When you think of a sample type you probably immediately envision a standard blood tube.
In 'the industry' we call these vacutainers (because there's a vacuum behind the seal that sucks the blood in…slurp!).
There's also lots of pretty colored tubes.
The one we like best for genetics is the lavender tube (purple top) because it's a tube that isn't full of potential inhibitors that might interfere with down-stream sample prep.
Yellow tops (ACD) and green tops (heparin) are also usually acceptable although heparin can be carried over as an interfering substance with some nucleic acid extraction methods (especially filter columns).
Streck tubes and PAXgene tubes are also pretty common for cell free DNA collection (non invasive screening) or applications that test RNA.
Saliva:
Spit tubes became really popular in genetic testing because of direct to consumer ancestry products!
Saliva is terrible in lots of ways, though.
It can be contaminated with all sorts of things found in the oral cavity.
Like, whatever was eaten last along with the bacteria that live inside your mouth.
Saliva samples can also be very viscous depending on the individual.
This makes them challenging to open without contaminating everything nearby but it also can make them very hard to pipette accurately!
Swabs:
Oral, anal, vaginal, nasal…basically any surface or orifice that can host an infectious agent or secrete something is a potential candidate for swabbing.
These come in two flavors, dry swabs (not ideal) or liquid stabilized (way better sample quality!).
For pediatric patients there are also now 'lollipops' to trick kids into thinking they're getting a treat when they're actually giving you a cheek swab!
Urine:
This one is a staple of the diagnostics industry, mainly for 'occupational testing' which is a euphemism for drug testing, but there are genetic screening applications that use urine too.
Some bladder and prostate cancer tests use urine as a sample type along with a number of PCR tests that use it to detect urinary tract infections.
One fun fact is that in occupational testing labs, the accessioners (scan samples in *beep*) note the 'aroma' of the sample.
You know, just in case it's actually apple juice.
💩💩💩:
At home collected fecal samples for microbiome or cancer screening are undoubtedly the worst.
They're like a box of chocolates, 'you never know what you're gonna get.'
Is it a bulk sample?
Did the patient follow the instructions?
Are there remnants on…everything?
But, before you try to answer ANY of those questions, make sure you have gloves on...
While chromosomes are usually depicted as X's, they actually spend most of their time jumbled up like a giant ball of yarn.
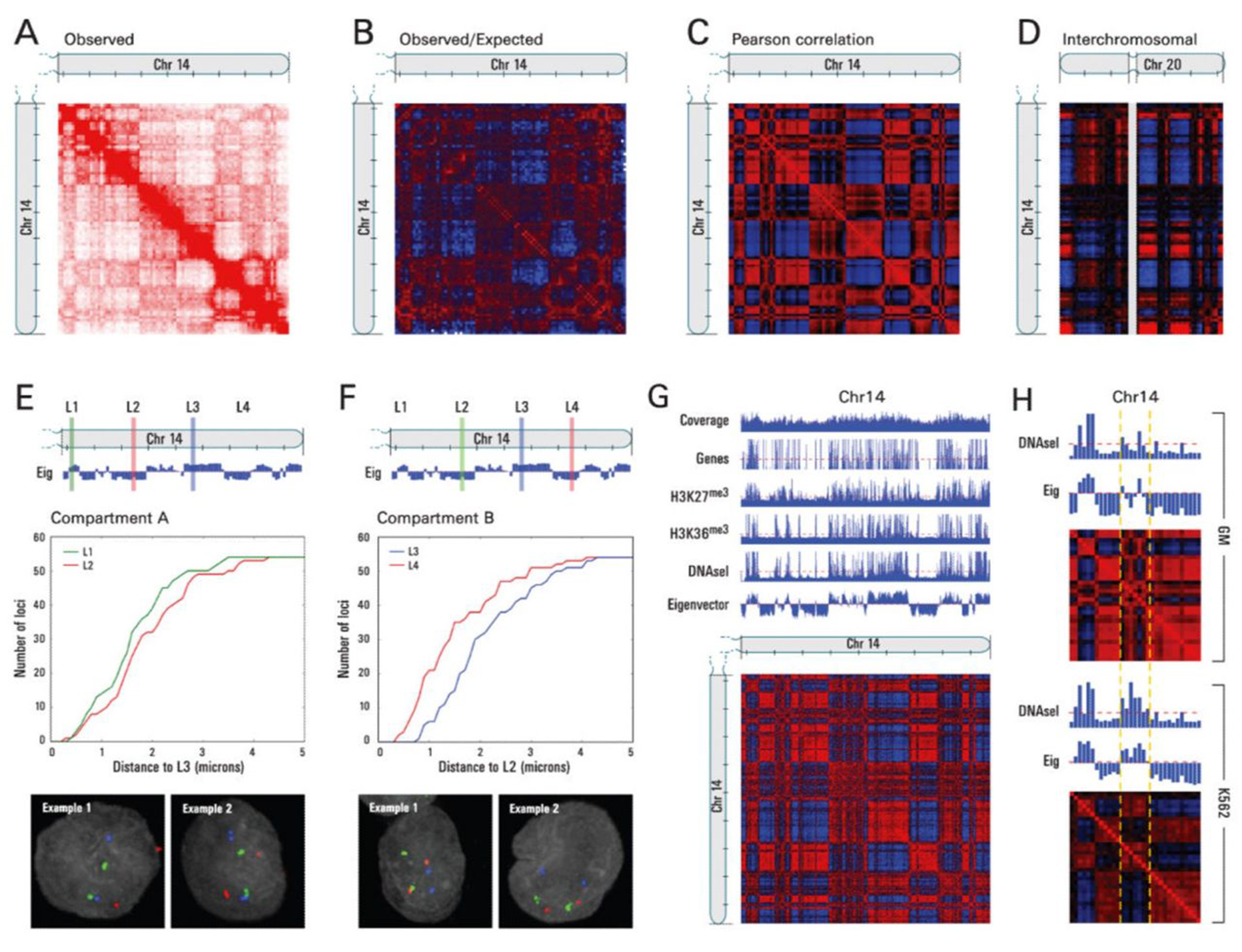
Friedrich Miescher first discovered nuclein (DNA) in the early 19th century, and determined it was bound to proteins.
But it was Walther Flemming in 1889 who suggested that this DNA-protein complex be called chromatin - which is the material that makes up our chromosomes.
Don’t be fooled, though.
Just because chromatin was described and named in the 1800’s, that doesn’t mean anyone actually understood what it did!
It took until 1950 for scientists to finally begin to understand that the ‘genetic’ material was DNA.
And it wasn’t until the 1960’s that we first started piecing together how chromatin actually functioned.
To this day, we’re still not entirely sure!
But the classical depiction of our genetic material as ‘rods’ or ‘X’ structures in many introductory biology textbooks is not totally accurate.
This is because these structures only appear when chromosomes ‘condense’ during cell division!
The vast majority of the time, most of our chromosomes are unwound in the nucleus and tangled together in a giant mass.
You might be wondering if the interweaving of our chromosomes in the nucleus plays an important role in regulating which genes are expressed?
And the answer to that question is:
You bet your chromatids it does!
While it had been hypothesized for many years that long-range interactions played an important role in gene expression, it wasn’t until 2009 that these chromosomal contacts were first characterized genome-wide.
Previous work using a technique called chromosome conformation capture or 3C had shown that regions of chromatin could form ‘loops’ or touch each other at long distances.
But, the authors here mapped chromosome contacts using a technique that they named Hi-C.
It differed from previous chromosome conformation capture methods in that it was the first to leverage high throughput sequencing to look at ALL of the connections within and between chromosomes to map the three-dimensional structure of the genome.
And what they found was really cool!
A) Shows a heat-map of interactions across chromosome 14, B) highlights areas of more contact than expected (red) and less contact than expected (blue), D) shows interchromosomal interactions between chromosome 14 and 20, E and F) highlight how linearly distant regions can actually be in very close proximity to one another, and G and H) show how this architecture correlates with gene rich, open and closed regions of chromosomes!
So, why are these results important?
Because they showed us how the physical three dimensional structure of our genome can impact gene expression, adding yet another regulatory layer to the story of epigenetics.
###
Lieberman-Aiden E et al. 2009. Comprehensive mapping of long range interactions reveals folding principles of the human genome. Science. DOI: 10.1126/science.1181369
Were you forwarded this newsletter?
LOVE IT.
If you liked what you read, consider signing up for your own subscription here:
