Omic.ly Weekly 31
June 30, 2024
Hey There!
Thanks for spending part of your Sunday with Omic.ly!
But, do you have a friend or a colleague who you think might also enjoy this newsletter?
Feel free to forward and make them fans of Omic.ly too!
This Week's Headlines
1) Why do women live longer than men? Sperm production is expensive
2) Genealogy databases aren't just for finding your relatives anymore
3) The 'Histone Code' fundamentally changed how we think about gene expression
Here's what you missed in this week's Premium Edition:
HOT TAKE: If the allegations are true, Illumina's acquisition of Grail appears to not just be a bad decision, but a case of fraudulent enrichment
Or if you already have a Premium Subscription:
Are you a guy? Do you want to live longer? It might be time to ditch your sperm.
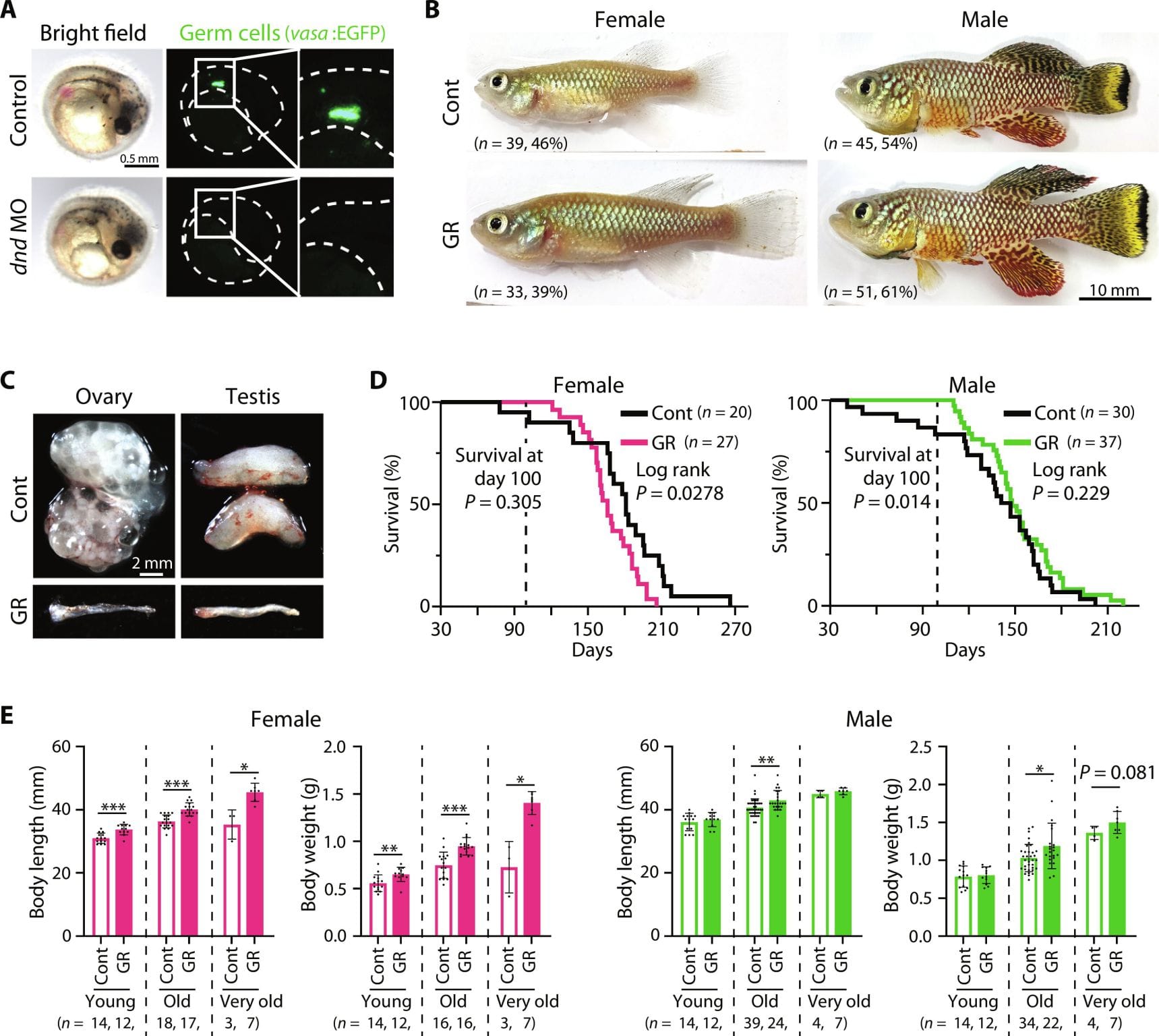
Females live longer than males.
It’s like a universal rule of the animal kingdom.
In humans, females outlast their male counterparts by about 5 years with women living until ~80 and men only reaching 75.
That’s about a 7% difference in life expectancy!
What explains this difference?
A number of factors have been hypothesized, such as males take more risks, are more prone to suicide, and women tend to lead much healthier lifestyles.
But that's just in humans!
This sex difference in life expectancy extends to many other animals.
And it’s thought that at least some of these life expectancy differences are a trade-off between an organism’s own needs and the constant production of sex cells known as gametes.
Studying these compromises among growth, development and reproduction throughout the life of an organism, or their Life History, can help explain the differences we see in animal size, age of maturation and their life expectancy.
But it has been hypothesized that the creation and maintenance of gametes is very expensive and experiments in non-vertebrates (fruit flies and worms) have confirmed that the molecular removal of sex cells can extend an organism’s life.
Similar work has not been done in vertebrates; however, an analysis of Korean eunuchs (males who have been castrated) from the 1800’s found that men who no longer had testicles lived between 14-19 years longer than the general population.
Castration seems a bit extreme, and there are other confounding factors that could account for the increased lifespan of these eunuchs, for example, they were very well taken care of as members of the royal staff.
And, at least in worms, complete removal of the gonads has no effect on their life expectancy.
So to settle this line of questioning once and for all (not really, because this was done in fish), the researchers behind today’s paper knocked out the gene responsible for creating gametes in both male and female Killifish.
What they found can be seen in the figure above a) Confirmation of gamete removal. b) No effect on gender coloration. c) The gonads are MUCH smaller. d) Life expectancy of females decreased, males increased. e) Quantification of the physical size differences
The researchers went on to show that the differences between the sexes post-gamete removal appear to be mostly dependent on endocrine signaling and metabolism.
But the reduction of female life expectancy was unexpected and the researchers attribute this to a significant reduction in estrogen production.
This work is important because it could provide new strategies for combating aging (in men), and offer new pathways for us to target to improve human health and longevity.
###
Abe K, et al. 2024. Sex-dependent regulation of vertebrate somatic growth and aging by germ cells. Science Advances. DOI: 10.1126/sciadv.adi162
The killer app for consumer genetics literally is to catch killers. Here's why:

You may have seen news reports about the brutal stabbing of 4 University of Idaho students while they slept a few years ago.
Not much was released about the case other than a description of the suspect's vehicle, and that it appeared that a few of the victims woke up during the attack and fought back before being killed.
These murders left the University of Idaho community on edge since a motive was hard for local authorities to ascertain - by all accounts, this was a random act of violence.
Fortunately, a suspect was arrested a month later, with a footnote that apprehension was aided by genealogy databases.
Now, you might ask why this is an important detail, but way back in 2018, it was shown that genetic databases, like GEDmatch, could be used to identify most white Europeans, meaning, if your DNA was left at a crime scene, that DNA could be matched with your name.
GEDmatch, at the time, had 1.2m users who had publicly shared their genotypes.
Today, Ancestry and 23andMe together hold genotypes for more than 30 million people.
That's bad news for criminals.
If you've committed a rape or a murder in the US, and there is DNA evidence, you're probably going to be caught.
This works because the technology that Ancestry and 23andMe use basically serves as a highly accurate fingerprint.
And if you remember from biology class, you share 50% of your DNA with your mother and father and slightly smaller percentages with your cousins and other extended family members.
But, contrary to popular belief, these companies do not perform genome sequencing.
They sample common mutations that are scattered across the human genome using genotyping arrays.
While your genome is composed of 3 billion base pairs, genotyping arrays look at about 700,000 locations.
And because you share many of these genetic differences with your relatives, they can be used to link you to your extended family and ancestors through genealogy databases.
That's the consumer use.
On the flip side, law enforcement can use these same databases to identify criminals, essentially by doing the same thing, submitting a sample to a database, and finding all of the relatives that are linked to that sample.
Add in a little bit of sleuthing, and it's pretty easy to narrow down a very short list of potential owners of the original sample.
From there it's just a matter of verifying the match.
The Golden State killer was famously caught after authorities grabbed his garbage from the curb for verification.
Other cases have been solved by compelling suspects to submit a blood or a hair sample for confirmation testing.
In the Idaho case, it was revealed that the killer was caught using 'touch' DNA found at the crime scene.
I'm thankful we have these tools available to put violent criminals behind bars.
It's only a matter of time before we catch them all.
Histones were once thought to just be dumb structural proteins that held chromatin together; now we know that the ‘histone code’ is a major regulator of gene expression.
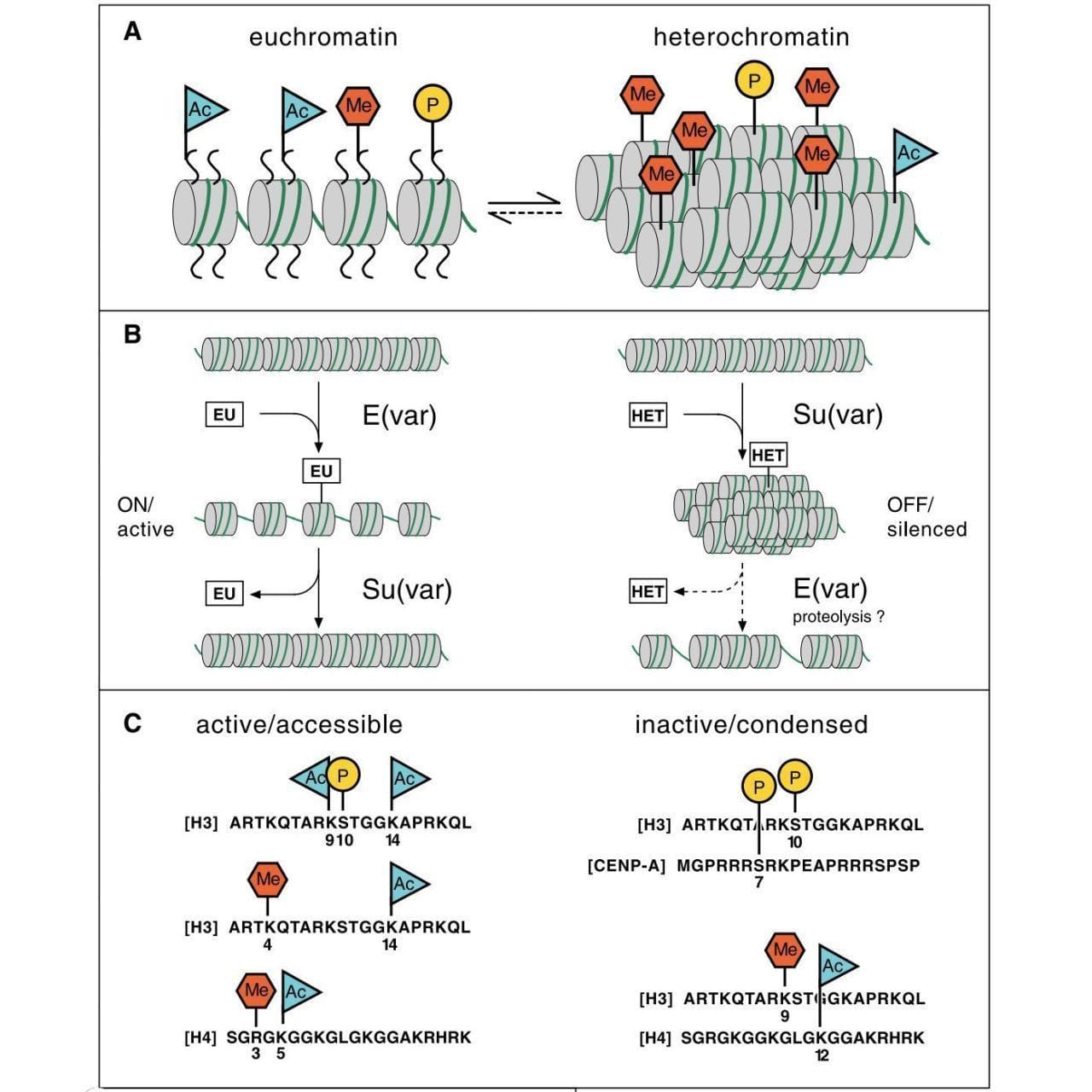
The first studies on gene expression were done in bacteria.
Here, gene expression is very straightforward:
Proteins bind to DNA, turn it into RNA and then that RNA gets read to create other proteins.
Things get much trickier in complex organisms.
Their larger genomes are packaged into a super-structure called chromatin.
And, in 1974, Roger Kornberg proposed that chromatin was made up of a DNA-protein complex that he named the ‘nucleosome.’
DNA is wrapped around histone proteins to create this highly ordered structure but this also presents an interesting problem to solve.
If DNA is all coiled up, how does the gene expression machinery get in there to keep cells alive?
Fortunately, David Allis suspected that histones were critical players in gene regulation.
He remembered earlier work from Vincent Allfrey showing that histones could be methylated or acetylated and he was determined to figure out if these modifications performed a function.
His chosen model for this was the single-celled protozoan Tetrahymena.
This might seem an odd choice, but Tetrahymena are full of acetylated histones and have high levels of gene expression which made them perfect for the task!
Allis and his team figured an enzyme was responsible for all of this acetylation and isolated the first histone acetyltransferase (HAT) from them in 1996.
They found that this enzyme, p55, was similar to Gcn5p, a protein known to regulate gene expression in yeast.
This discovery, along with additional work from Michael Grunstein, connected HATs to gene activation.
But adding an activation mark to histones only solves half of the gene expression problem.
How do you turn genes off?
Fortunately, the first histone deacetylase (removes acetyl groups) was discovered soon after by Stuart Schreiber and this explained how acetylation levels were maintained in cells.
Following these two discoveries, additional histone modifying enzymes were found that added phosphorylation and methylation to the mix.
The figure above pulls all of this together into what is known as the histone code, or, how histone modifications are added, read, and erased to control what regions of DNA are accessible for gene expression.
A) displays the histone marks associated with open (euchromatin) and closed (heterochromatin), B) shows a model of how suppressors and enhancers of variegation work to open and close chromatin, and C) shows which amino acids on each histone are marked in open and closed chromatin.
But, this seminal article is important because it is one of the first to link histone modifications to epigenetics and this idea fundamentally changed how we viewed gene regulation at the chromatin level, forever.
###
Jenuwein T, Allis CD. 2001. Translating the histone code. Science. DOI: 10.1126/science.1063127
Were you forwarded this newsletter?
LOVE IT.
If you liked what you read, consider signing up for your own subscription here:
Subscribe to Omic.ly
