Omic.ly Weekly 30
June 23, 2024
Hey There!
Thanks for spending part of your Sunday with Omic.ly!
This Week's Headlines
1) A new genetic cause for Inflammatory Bowel Disease has been discovered
2) A trip down the Omics rabbit hole
3) mRNA vaccines almost weren't a thing
Here's what you missed in this week's Premium Edition:
HOT TAKE: The temperature has gone from hot to cold post-IPO for Tempus, but have no fear, they added AI to their name
Or if you already have a Premium Subscription you can
Inflammatory diseases affect 5% of the population but we have a poor understanding of how to treat them. Multi-omic studies are changing that.
Inflammatory and autoimmune diseases like lupus, psoriasis, Crohn’s disease and ulcerative colitis can be severely debilitating.
With respect to the last two, which are inflammatory bowel diseases (IBDs), symptoms can include rectal bleeding, severe abdominal pain, and diarrhea.
These conditions are usually diagnosed by looking at inflammatory markers in stool samples or viewing inflamed tissues via colonoscopy or endoscopy.
The exact cause of the inflammation in IBD is not well known, although flares in some people can be associated with eating habits or disruptions of the microbiome.
This has had the effect of confusing people about the difference between it and irritable bowel syndrome (IBS) which can have similar symptoms but whose cause is more related to infections (like C․diff), bacterial overgrowth, or other changes that affect gut motility.
But because IBD is associated with inflammation, and co-occurs frequently with other autoimmune diseases, it’s hypothesized that there is a strong genetic component to the disease.
Genome wide association studies (GWAS) have identified over 200 regions of the genome linked to IBD; however, only a few of these regions have mechanistic associations with inflammatory genes.
Unfortunately, many of these variants fall in non-coding regions or gene deserts and so it is thought (as is true for much of GWAS) that these associated regions are involved in epigenetic regulation of gene expression in affected cell types.
To that end, the researchers behind this week’s paper chose to study a region of the genome associated with multiple inflammatory diseases, chromosome 21q22 (chr21q22).
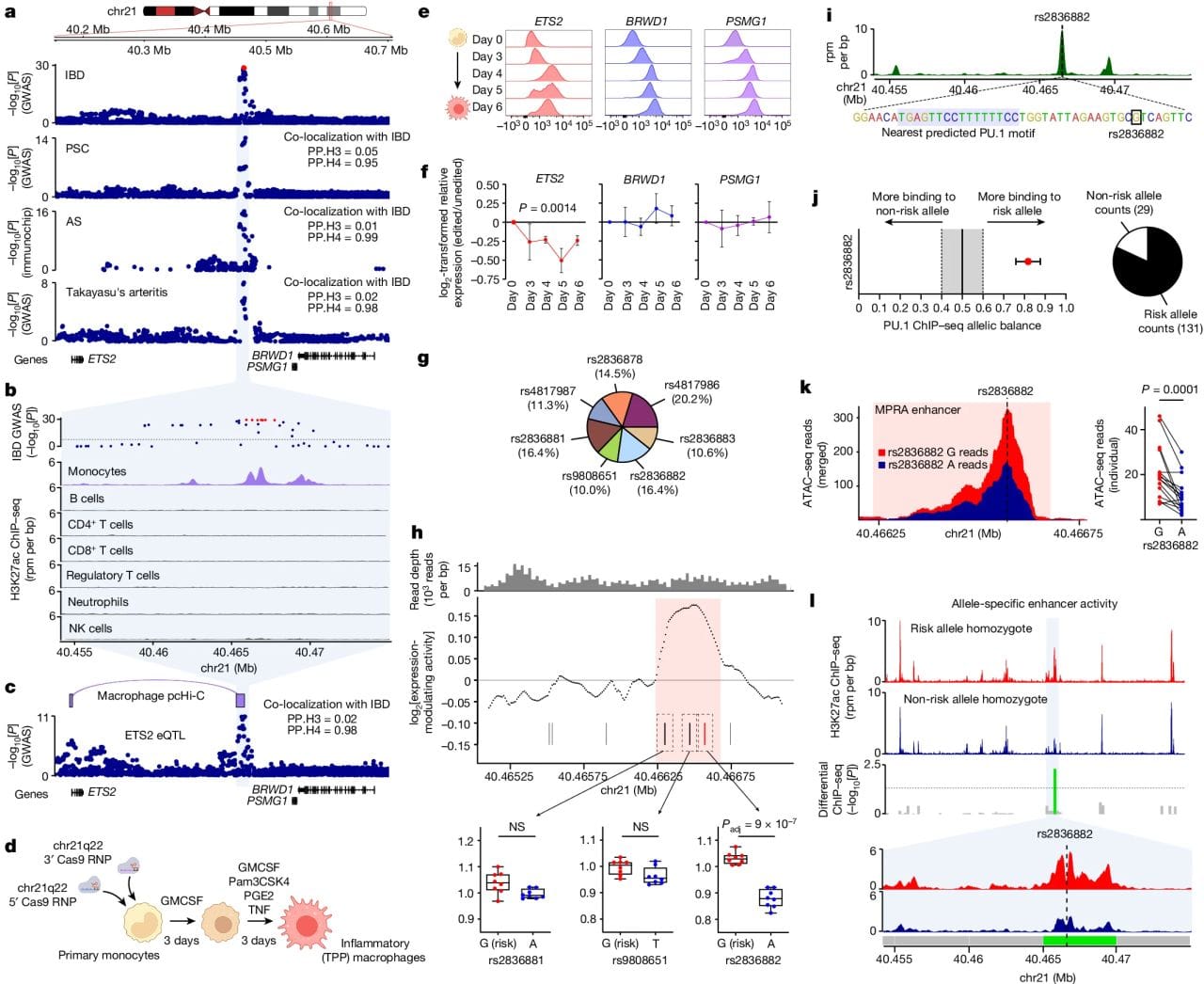
In the figure above you can see a) the GWAS association of this region with IBD, primary sclerosing cholangitis (PSC), ankylosing spondylitis (AS) and Takayasu’s arteritis. b) this region is associated with activating H3K27ac epigenetic marks only in inflammatory monocytes. c) this region is physically linked to the ETS2 gene, a transcription factor. d, e, f) CRISPR removal of this region results in downregulation of ETS2 expression. g,h,i,j,k,l) a specific SNP, rs2836882, is associated with recruiting the chromatin remodeling protein PU.1 to this newly discovered ETS2 enhancer.
The authors go on to show that CRISPR–Cas9 disruption of ETS2 reduced pro-inflammatory markers and single-cell transcriptomics of diseased IBD tissue confirmed the presence of higher levels of ETS2 in associated monocytes.
While more work needs to be done, it appears that targeting the ETS2 pathway presents a promising therapeutic strategy for treating conditions like IBD.
###
Stankey CT, et al. 2024. A disease-associated gene desert directs macrophage inflammation through ETS2. Nature. DOI: 10.1038/s41586-024-07501-1
What’s in an 'ome?' Basically everything.

Naming an 'ome' was very popular in the late 90's and early 2000's, probably because that's right around when we all caught the 'Genome' bug.
But the majority of these 'omes' are derivatives or sub-omes of the major ones.
Some of these may only still exist in the mind of their creator, and don't get mad at me if I missed your favorite one - there are a lot!
What follows is a long list of the ones I've heard of before:
Antibodyome - All the antibodies in an organism
Cellome - The totality of molecules and their interactions within a cell
Chemome - The totality of chemicals in cells or organisms
Cytome - All the cells of a particular organism together with their associated cellular processes
Degradome - All the compounds produced in the degradation of a material (especially of a protease)
Embryome - All the cell types which arise during embryogenesis and development
Exome - The complete exon content of an organism or individual; the subset of the genome that excludes introns
Exposome - All of the effects of life-long environmental exposures on health
Glycome - All the polysaccharides, glycosides and carbohydrate-related compounds in an organism
Immunome - All the genes and proteins associated with an immune system
Interactome - All the interactions between biological entities in a cell or organism
Kinome - All the kinases expressed in a cell or contained in a genome
Lipidome - All the lipids in a cell or organism
Ligandome - All the molecular ligands for proteins in a cell or organism
Methylome - All the nucleic acid methylation modifications in an organism's genome
Microbiome - The community of microbes in or on an organism
Neurome - The totality of the neurons in an organism
Nosolome - The totality of diseases and their association network.
ORFeome - The totality of open reading frames in a genome
Pangenome - The genome of all the strains of a particular species
Paranome - The complete set of paralogous genes in a genome
Pharmacogenome - All the genes that are affected by pharmaceutical drugs
Phenome - The whole set of phenotypic entities in an organism
Regulome - The whole set of regulation components in an organism, usually used in the context of signal transduction
Reactome - All the biochemical reactions that take place within an organism
Secretome - All of proteins secreted from a cell
Signalome - All the signalling pathways of an organism or cell
Supergenome - All the genes encoded on an organism's chromosomes together with those encoded on mobile genetic elements
Toponome - The spatial network code of proteins in cells and tissues
Toxicome - The totality of toxic chemicals in cells
Ubiquitome - All the ubiquitin polypeptides that play a part in modifying and degrading proteins
There's even an 'ome' for people who hate the propagation of superfluous omes…the Antiome.
You can count me as a member.
Sorry, Padme.
It is estimated that COVID-19 vaccination has prevented over 20 million deaths worldwide. We can thank the dogged perseverance of a browbeaten scientist for that.
Katalin Karikó’s love of mRNA began during her postdoc at the Biological Research Centre of Hungary.
She had a feeling mRNA was destined for greatness, and moved to the US with her family in 1985 to continue her pursuit of turning it into a therapeutic.
However, as a foreign scientist in the US, she was often passed over for faculty positions and grants.
She persisted in spite of these setbacks.
Fortunately, a chance meeting with Drew Weissman led to one of the most important discoveries in modern medicine.
In 1997, Weissman had just completed his fellowship under the supervision of Anthony Fauci, and was in desperate need of some mRNA.
Karikó provided that mRNA but they noticed that it caused an inflammatory response in mice, which, unfortunately, would limit its usefulness as a therapeutic.
Undeterred, the pair decided to figure out why.
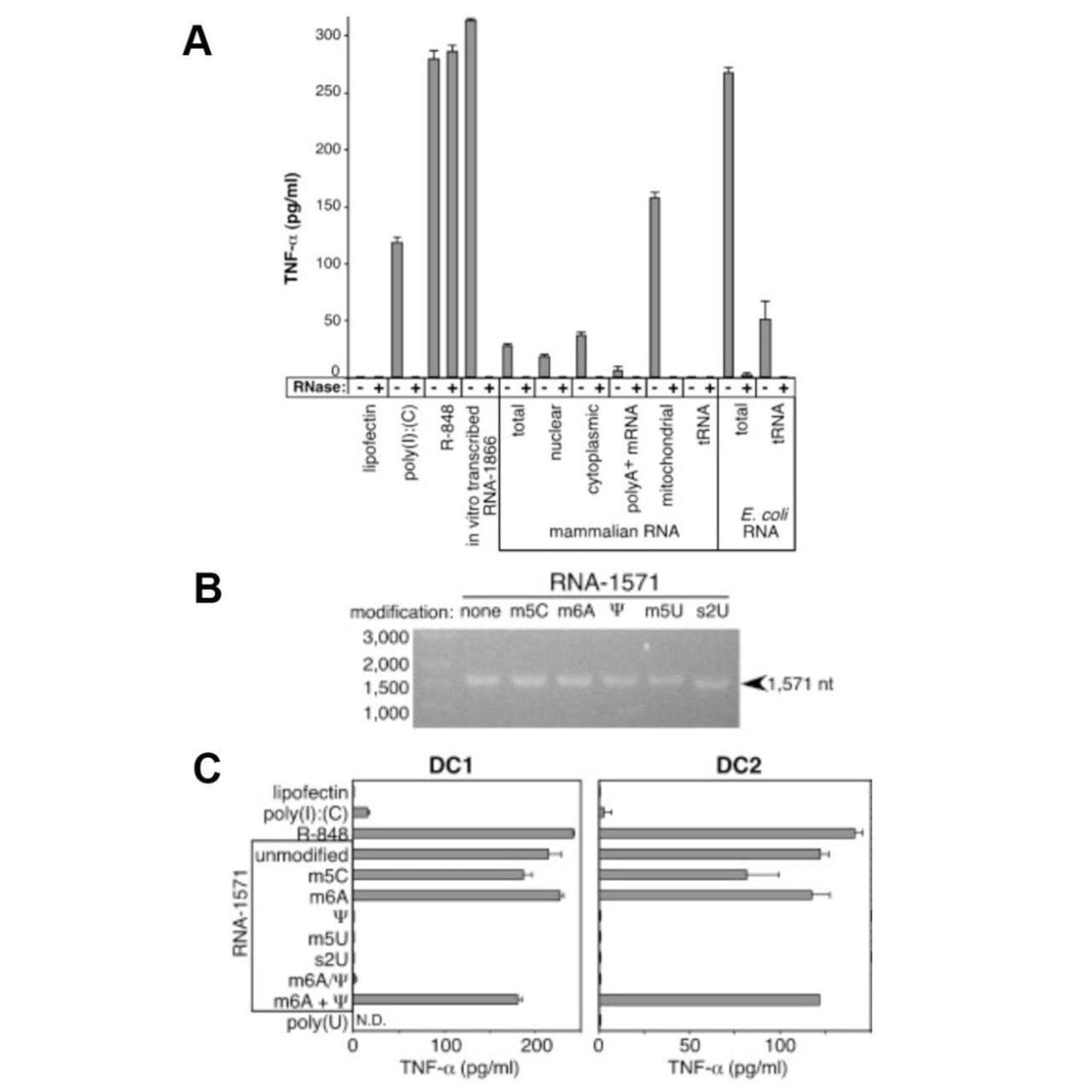
The figure above shows what they discovered.
In A, Dendritic Cells (produce TNF-α, an immune cytokine) were treated with 3 controls (poly I:C - synthetic RNA, R-848 - a drug, and in vitro transcribed RNA) which are known to stimulate the immune system along with isolated mammalian RNAs, and bacterial RNAs.
Only mitochondrial RNA and bacterial RNA induced a strong response (in addition to the controls), while tRNA had no effect!
So, what accounts for this difference?
Karikó and crew thought that since 25% of tRNA bases are modified, maybe that’s what prevented most of the mammalian RNAs from sounding the alarm!
In B, mRNA was transcribed using modified nucleotides: 5-methylcytidine (m5C), N6-methyladenosine (m6A), pseudouridine (Ψ), 5-methyluridine (m5U), or 2-thiouridine (s2U).
And in C, those mRNAs were introduced into dendritic cells.
The modified uridine containing mRNAs failed to cause an immune response.
Success!
Karikó later showed that Ψ containing mRNA produces the most protein and that those transcripts last for up to a month.
However, neither the institution where she was employed nor the US government recognized the tremendous potential of her work.
And so, she left the 'Mean Girls' culture of US academia and joined a little German company called BioNTech.
She continued her collaboration with Weissman and, with BioNTech, developed the first experimental vaccine using modified mRNAs against Zika in 2017.
BioNTech went on to produce the first mRNA vaccine for SARS-CoV-2 with doses being delivered worldwide in less than 8 months.
The recent value of this achievement can be measured in tens of millions of lives, but its future has the potential to save hundreds of millions more.
Karikó and Weissman were awarded the Nobel Prize for this work in 2023.
###
Karikó K et al. 2005. Suppression of RNA Recognition by Toll-like Receptors: The Impact of Nucleoside Modification… . Immunity. DOI: 10.1016/j․immuni.2005.06.008
Were you forwarded this newsletter?
LOVE IT.
If you liked what you read, consider signing up for your own subscription here:
