Omic.ly Weekly 29
June 16, 2024
Hey There!
Thanks for spending part of your Sunday with Omic.ly!
This Week's Headlines
1) Have you ever wanted a superpower? Me too!
2) Metabolomics: Hype or Heroine?
3) The 2-D structure of tRNA was solved in 1965, its solver probably isn't someone you've heard of before
What you missed in this week's Premium Edition:
HOT TAKE: If you thought the sequencing wars were hot already, have you considered what a $10 genome might do to them?
Or if you already have a premium subscription:
Some human populations have genes that would even make Poseidon jealous!
Humans inhabit a wide variety of environments.
We can be found in extremely cold places, like the arctic, and extremely hot places, like the Sahara desert.
Some, like the people of Tibet, have settled at high altitudes, while others, like the ancestors of the Bajau ethnic group in southeast asia, spent much of their time at sea diving for food.
And unsurprisingly, each of these populations of humans have acquired unique genetic adaptations that helped their ancestors (and them!) thrive in their environments.
People living in cold places have higher metabolisms to generate more heat while people who live in desert environments sweat more to keep themselves cool.
Similarly, Tibetans have genetic changes that increase the number of oxygen carrying red cells in their blood and allow them to live at high (low oxygen) altitudes.
The Bajau of southeast Asia are also uniquely adapted and are well known for their ability to free dive for extended periods of time.
While most Bajau now live a more terrestrial lifestyle, there are still some tribes that spend the majority of their lives on the water.
And for some of them, this means diving and hunting for food for 5 hours every day!
But you might be thinking, “I shiver when I'm cold, if I go to high altitude, the number of red cells in my blood increases, and if I practice, I can hold my breath for a long time too!”
Which is a reasonable observation, and training certainly plays a role here, but these populations can do all of these things better, faster and longer.
And in the case of the Bajau, and other cultures who dive for extended periods of time, we already know they possess distinct physiological changes that make this possible.
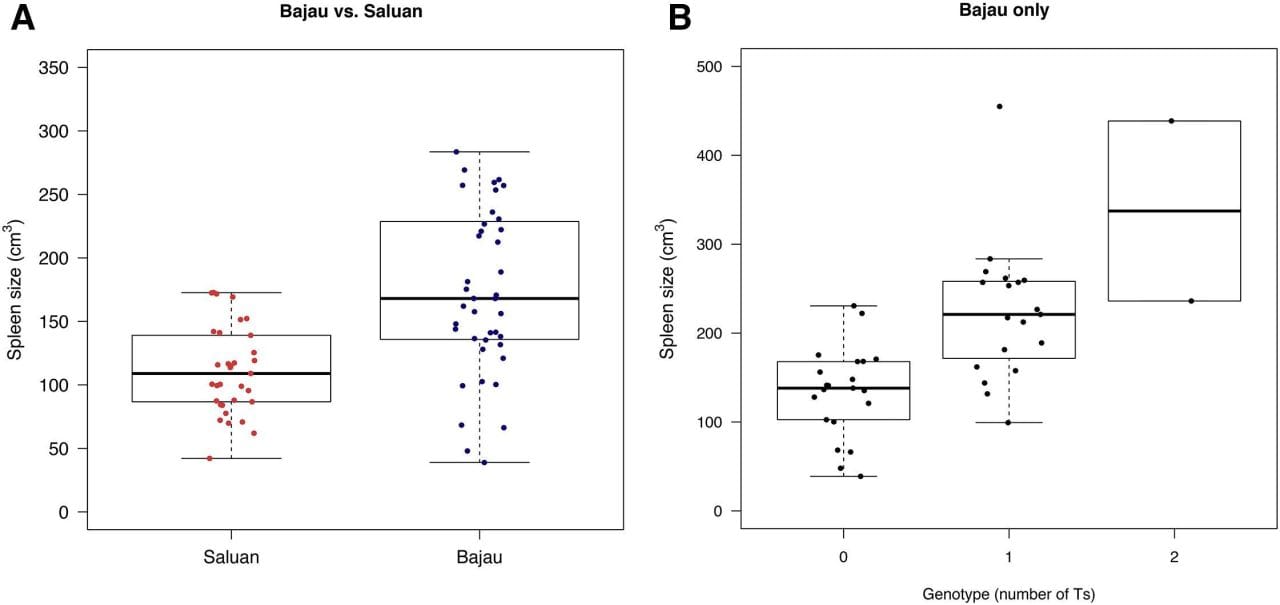
One of these is that the Bajau have abnormally large spleens which are used to store extra red blood cells that can be released during diving.
But the genes associated with the exceptional diving capabilities of the Bajau are largely unknown.
To change this, the researchers behind today’s paper used genetic association studies to find a few candidates and they correlated one of them, PDE10A, to increased spleen size in the Bajau population (See the figure above).
Mutations in PDE10A have been shown to be associated with hyperthyroidism and increased spleen size in other populations.
But this research also identified changes in three other diving associated genes: FAM178B (helps maintain blood pH in the presence of high CO2), BDKRB2 (limits oxygen use), and CACNA1A (releases the neurotransmitter glutamate in response to low oxygen).
These findings highlight the important roles that the environment and human culture can play in shaping our genomes.
And how these pressures have given some humans superhero capabilities!
###
Ilardo MA, et al. 2024. Physiological and Genetic Adaptations to Diving in Sea Nomads. Cell. DOI: 10.1016/j․cell.2018.03.054
How does the promise of metabolomics stack up against our current reality? It’s complicated.

One of the great enduring mysteries in biology is how we go from the DNA in our genome to a functional organism.
The simple explanation is that the genome codes for RNAs that code for proteins and those proteins make up our cellular structures or they synthesize those things from raw materials.
'But I thought this was a post about the metabolome, Brian?'
It is!
All of those raw materials, the leftovers, and the waste are called metabolites and it's metabolites that make up the metabolome!
So, in essence, the metabolome is all of the substrates, intermediates and end products of cellular metabolism.
But how does the metabolome help us figure out how the genome creates a functioning human?
Because the signatures of the metabolome actually tell us quite a bit about what proteins the genome has expressed AND what those proteins are doing at any point in time!
Every cell, tissue, and body fluid have their own metabolome and these can change over time as a part of development, aging, or even environmental factors.
Since the metabolome represents all of those cellular activities, it also can be used in a variety of important ways to track health and disease.
There are 2 key clinical uses for metabolomics and one more aspirational use case:
Drug Dosing and Drug Metabolism - Patients and physicians struggle to find the correct therapies or the correct doses of those therapies. This unfortunately becomes an individualized trial and error process. With metabolic profiling we can actually see how the body deals with a drug. Is it processed quickly or does it cause any effect at all? We can use this information on how the body metabolizes a drug to zero in on dosing and response much more quickly than we can by the 'take 3 of these and call me in the morning' method.
Metabolic Disease Diagnosis and Monitoring - metabolic diseases are some of the hardest to diagnose because they often 'look' like other things. This is especially bad if someone is misdiagnosed and placed on an ineffective treatment. This means people can wrongfully be prescribed steroids, or immunosuppressants, etc when the real solution could be a change in diet or supplementation with something their body is improperly metabolizing.
Health and Wellness - generalized screening for health and disease is where all of this gets a little tricky because the metabolic signals are less obvious as they relate to health and non-metabolic disease. This is a bit of a conundrum because to pull out any useful trends you need a lot of data, but what do you tell people in the meantime while you figure out the relevant signatures?
Which means that for widespread use outside of therapeutics and metabolic diseases, there's a bit of work to do to before anyone can provide actionable information to healthy populations.
The next great mystery to solve after the discovery of the DNA double helix was to figure out how the nucleotide sequence coded for proteins.
This was a challenging concept in the 1950's because prior to the publication of the Hershey-Chase experiment in 1952, there was a dispute over what the genetic material actually was.
Linus Pauling and other prominent scientists believed that the genetic material was protein.
They reasoned that since there were 20 amino acids, protein was obviously the genetic material because DNA only had 4 nucleotides and lacked the complexity to store information!
However, Alfred Hershey and Martha Chase showed with the help of a blender full of bacteria (and phages) that DNA, and not protein, was unequivocally the genetic material.
This presented a significant problem.
Watson and Crick get credit for solving the structure of DNA in 1953 (using data from Rosalind Franklin), but Crick's later theoretical work postulating how 4 nucleotides could code for all 20 of the amino acids was just as important.
In a lecture in 1957, he laid out 3 hypotheses (and the central dogma!):
1) RNA is the intermediary between DNA and protein
2) Certain RNAs serve as amino acid adaptors
3) These 'adaptor RNAs' use a triplet code to translate the genetic information into protein
Concurrently and without knowledge of Crick's hypotheses, Paul Zamecnik and Mahlon Hoagland discovered that small, soluble, RNAs could be charged with amino acids through an enzymatic process in 1958.
This independently confirmed Crick's adaptor hypothesis.
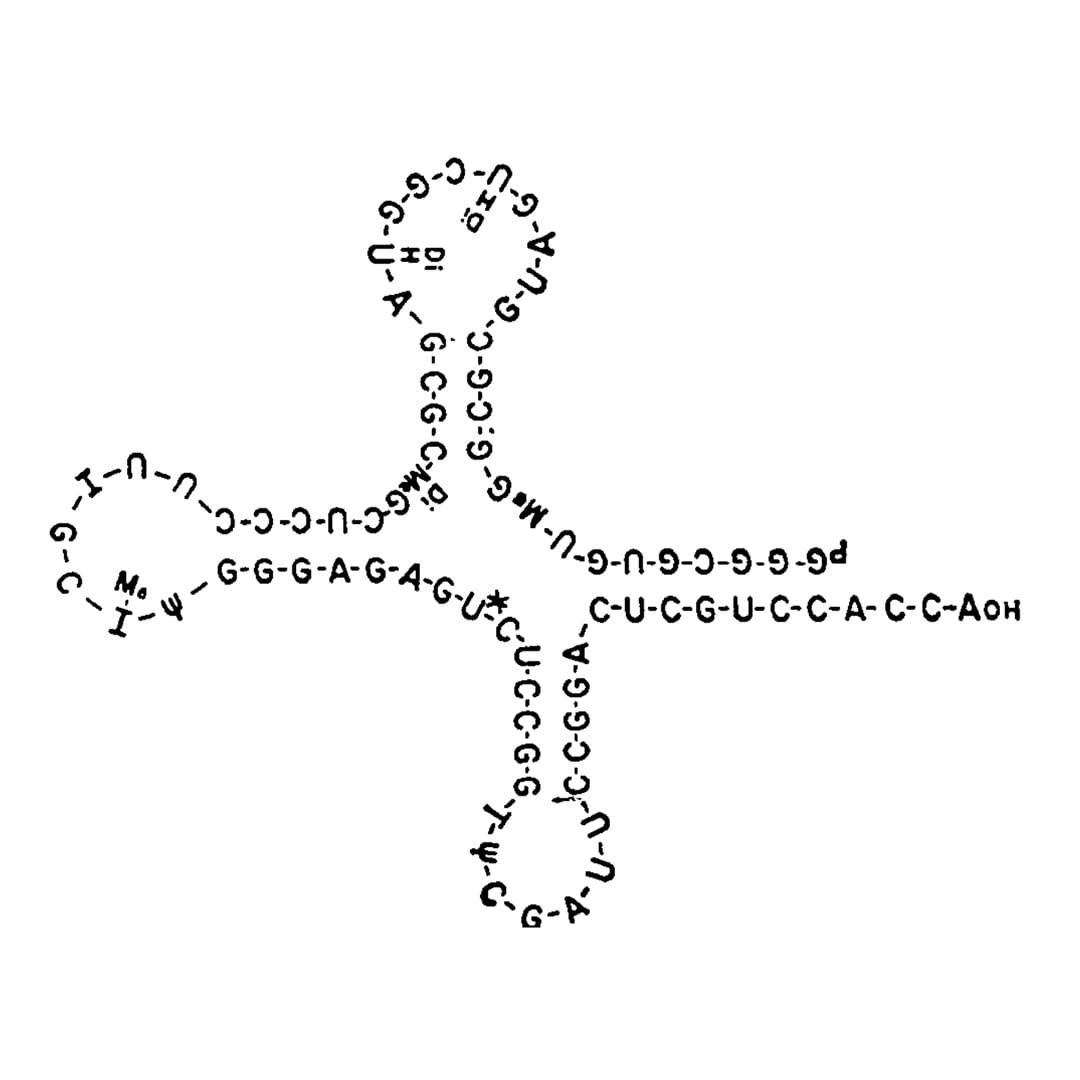
However, it was Robert Holley and his team at Cornell that determined the structure of this amino acid charged RNA which we now refer to as transfer RNA or tRNA.
Holley and team spent 9 years methodically sequencing tRNA using pancreatic and Taka-Diastase ribonucleases to chop up the RNAs into small fragments and then piece them back together.
They published the complete structure of all 77 nucleotides of tRNA-ala in 1965.
Ultimately, its the structure of tRNA that explains how it functions in the translation of RNA.
Holley shared the Nobel Prize with Khorana and Nirenberg, who deciphered the triplet code, in 1968.
The Nobel winning cloverleaf structure of tRNA depicted in that 1965 publication can be seen above.
It was proposed by a member of Holley's team, Elizabeth Beach Keller.
She was acknowledged for 'helpful suggestions.'
While her name does not appear as an author nor is she credited in textbooks that contain this now famous structure, her 1997 obituary tells the story of how she modeled tRNA using pipe cleaners, paper and velcro.
She sent the final structure to Holley in a Christmas card.
Her friend, Joseph Calvo, remembered her work during that time, "Her contributions were always behind the scene, looking until she found the shape that something wanted to take.''
###
Holley RW et al. 1965. Structure of a Ribonucleic Acid. Science. DOI:10.1126/science.147.3664.1462
Were you forwarded this newsletter?
LOVE IT.
If you liked what you read, consider signing up for your own subscription here:
