Omic.ly Weekly 28
June 9, 2024
Hey There!
Thanks for spending part of your Sunday with Omic.ly!
But, don't forget!
You can peruse all of these articles on the website throughout the week at your leisure if you don't have time to read everything today.
This week's headlines include:
1) Coming to a dermatologist near you: The microscope will see you now.
2) Make way for the METABOLOME ..ome ..ome.. ome!!!
3) Mice, Frogs, Rabbits, OH MY! The early days of pregnancy testing were wild.
What you missed in this week's Premium Edition:
HOT TAKE: ONT's ElysION is the most interesting thing happening in sequencing right now
Or if you already have a premium subscription:
We've heard a lot about liquid biopsies, but what if I told you non-invasive virtual ones are now a possibility?
Tissue biopsies are one of the most common diagnostic procedures.
If you’ve ever had a weird mole and gone to a dermatologist, they’ve probably cut a piece of it off (biopsy!) and sent it to a pathologist to figure out if it’s cancerous.
Biopsies are always invasive which is undesirable because they can be painful or leave scars even for benign things.
But what happens after the biopsy is taken is also pretty undesirable.
A pathologist doesn’t just put that chunk of your tissue under a microscope and shout, “Eureka, cancer!”
There’s a ton of preparation that happens before that!
The biopsy is embedded in paraffin, sectioned on a microtome (cuts insanely thin slices), stained with hematoxylin and eosin (H&E) and THEN it’s viewed by a pathologist.
This process can take days and it’s a highly skilled affair.
Wouldn’t it be easier if we could just peer into our skin directly with a microscope without all of the cutting, slicing and staining?
Of course it would, and that’s what the authors of today’s paper set out to do.
Microscopy techniques have advanced significantly over the past few decades and we can now see through multiple layers of skin.
These techniques include optical coherence tomography (OCT) or reflectance confocal microscopy (RCM), but interpreting images generated with these techniques requires training and they still have lower resolution than more traditional tissue section based microscopy.
To fix these problems, the authors used AI and machine learning to generate OCT images that looked exactly like their more labor intensive H&E counterparts.
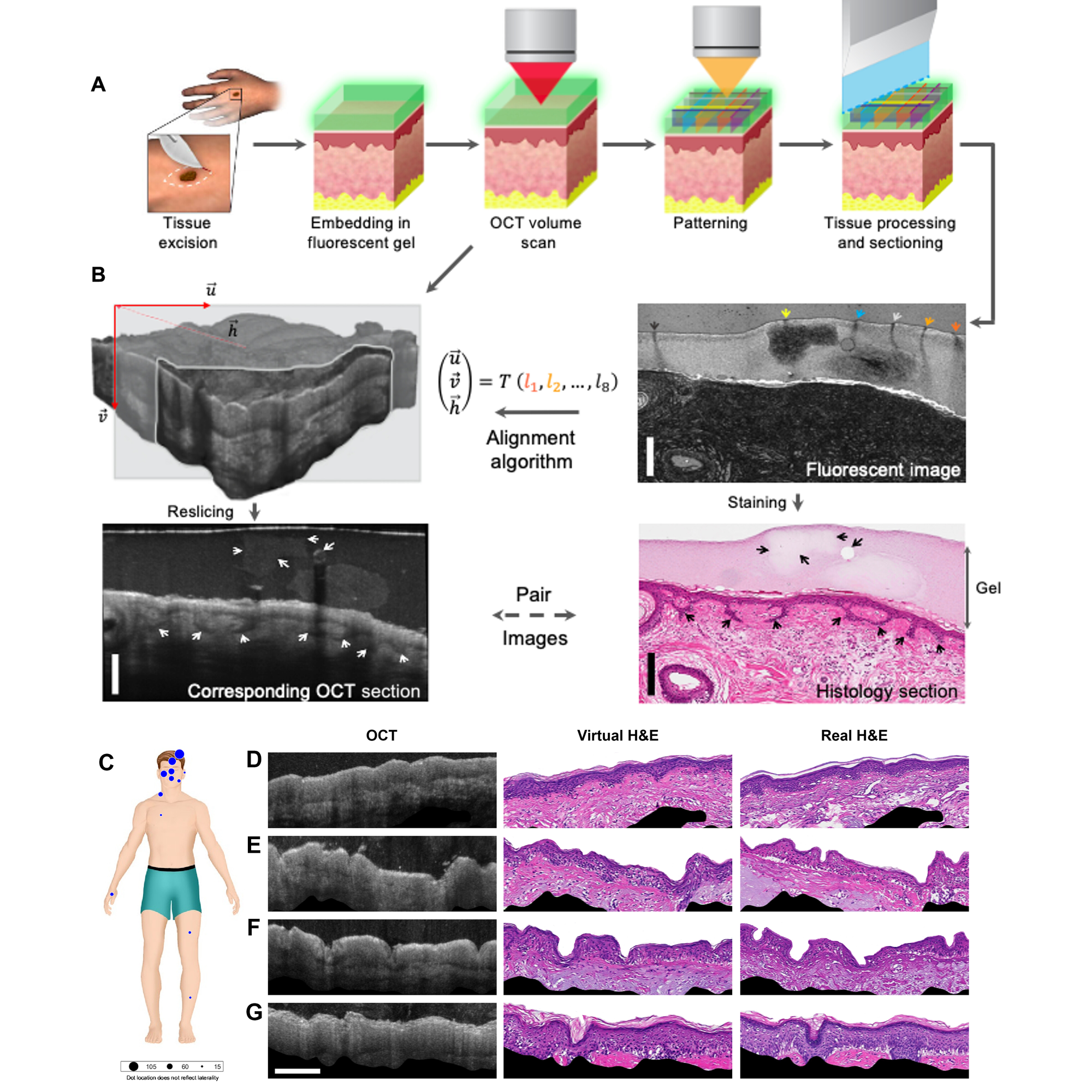
This work is detailed in the figure above.
In A) a traditional biopsy was taken, covered in a gel packed with fluorescent beads, scanned to make OCT sections and find the beads, and then the sample was sectioned and imaged normally.
This process generated the AI training data set seen in B) where OCT images with fluorescent bead markers were used to coregister (fancy word for lining up) them with the real H&E stained sections.
Finally, the trained algorithm was then used to generate virtual H&E stained sections (D-G) from OCT images of multiple skin biopsies (C).
While not perfect, the virtual biopsies were able to correctly ‘stain’ and identify complex structures like hair follicles with a resolution of 59 μm.
Further refinement of the technique with better training data and algorithmic optimizations are still required.
But a future that includes true virtual biopsies seems close, and could be key in enhancing our noninvasive diagnostic capabilities.
They'll also make trips to the dermatologist much more enjoyable!
###
Winetraub Y, et al. 2024. Noninvasive virtual biopsy using micro-registered optical coherence tomography (OCT) in human subjects. Science Advances. DOI: 10.1126/sciadv.adi5794
What's the biggest Ome of them all?

The METABOLOME ..ome ..ome.. ome!!!
There are a lot of Omes:
The Genome - stores all of our genetic information in two sets of 23 chromosomes.
The Epigenome - the non-DNA sequence alterations that impact gene expression.
The Transcriptome - all of the RNA transcripts that are produced as a result of gene expression.
The Proteome - all of the proteins that are produced as a result of RNA translation.
The Metabolome - all of the metabolites produced as a part of cellular metabolism.
Unfortunately, it's hard to study the metabolome because each successive 'ome' away from the genome represents an amplification.
One gene sequence in your DNA can produce hundreds of RNA transcripts, which in turn can be translated into thousands of copies of a protein, which then can produce tens of thousands of metabolites as a result of performing a cellular function.
There might be 22,000 genes in a genome but there are billions of metabolite molecules in a metabolome!
But what exactly is a metabolite?
It's defined as a substrate, intermediate or an end product of cellular metabolism. So they cover the whole range of metabolic inputs and outputs.
We know that metabolites can function as signaling molecules and serve as important markers of disease or cellular function.
Recent improvements in high throughput analytical techniques have allowed us to study the metabolome more closely and create metabolic profiles.
There's still a lot to learn but we've found that each tissue, organ, body fluid and disease state can have their own unique metabolomes.
Studying how these profiles change over time is important for understanding health and disease because the metabolome is a direct read out of the physiology of an organism.
Part of the challenge of studying the genome, the transcriptome and the proteome is that it's hard to tell what the immediate functional impact any of these have on an organism.
Is a gene expressed? We have to look at the transcriptome. Does an RNA make a protein? We have to look at the proteome. Is a protein active? We have to look at the metabolome.
These Omes are interconnected in important ways and so we kind of have to look at all of them.
But, by looking at the metabolites present within a sample, we can basically work our way backwards through the Omes because metabolites are the ultimate end point of cellular function!
So how do we look at billions of metabolites at a time and then make sense of everything that we see?
The two most common methods here are mass spectrometry and nuclear magnetic resonance spectrometry.
Deciding what's signal and what's noise is the biggest challenge to date and we're still in the process of turning our bulk measurements into broadly applicable and actionable information.
But as with the other Omes, the metabolome represents an exciting area of continued research and is one of the Omes to keep an eye on!
For the better part of 40 years, pregnancy tests were done by injecting urine into animals and seeing what happened. No, seriously!
Prior to 1928, determining whether someone was pregnant was a waiting game.
However, that didn’t stop people from trying to predict whether a missed period meant that a baby was on the way.
These early 'tests' ranged from mundane to insane and usually involved 'analyzing' urine.
The reason for this is probably due to its proximity to the birth canal.
And purveyors of these early tests were often referred to as 'piss prophets.'
This hints at the accuracy of such methods, but urine was actually the ideal sample type!
Before we get to WHY that is, we need to talk about the birds and the bees.
We know now that one of the first hormones to appear after sperm fertilizes an egg is human chorionic gonadotropin (hCG).
It is produced by the trophoblast cells that surround a developing embryo which eventually go on to form the placenta.
hCG’s function is to signal the ovaries, specifically the corpus luteum, to continue producing progesterone and estrogen.
This prevents the sloughing off of the endometrium and its discharge which marks the end of the menstrual cycle.
It’s these hormones that serve as the basis for modern pregnancy testing.
But in the early 1900’s, ‘hormones’ were a totally new concept with the word itself being coined by Ernst Starling in 1905.
By the 1920’s, scientists were finally starting to unravel the secrets of how hormones regulate the differences between the biological sexes.
Early work by Selmar Aschheim and Bernhard Zondek showed that extracts from the anteropituitary (the back of the pituitary gland) could promote ovary development and growth in animal models.
In 1927, they discovered that the urine of pregnant mice contained many of these same hormones and it had a similar stimulatory effect on their ovaries.
They quickly realized that injecting human urine into mice and seeing if their ovaries grew could be used to diagnose pregnancy and published on this in 1928.
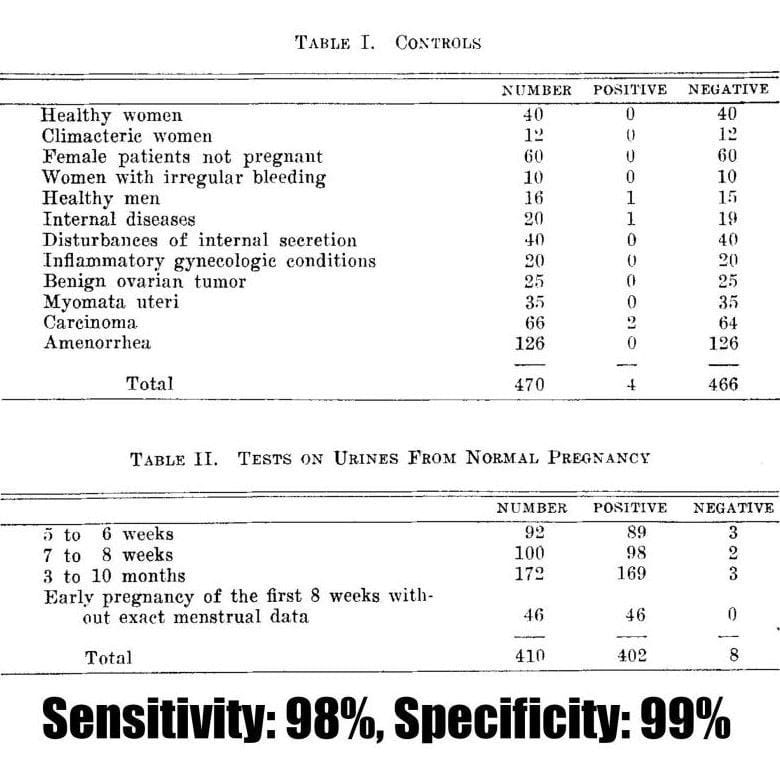
Unfortunately, this paper has no figures, but Ascheim published a follow-up paper in 1930 detailing the utility of the Ascheim-Zondek test for the diagnosis of pregnancy and placental tumors.
The two tables above display the impressive performance of the test in detecting 98% of pregnancies!
Further improvements came in 1936 with the Friedman or 'Rabbit' test in the US that gave results in 24hr, along with the development of a version using frogs that was popularized in the UK.
Animal based testing finally stopped in the 1960's when Leif Wide and Carl Gemzellthe invented an immunoassay to detect hCG directly.
However, the biggest innovation came in 1978 when at-home pregnancy tests first became available over the counter.
###
Aschheim S. 1930. The early diagnosis of pregnancy, chorionepithelioma and hydatidiform mole by the Aschheim-Zondek test. Amer J Ob Gyn. DOI:10.1016/s0002-9378(30)90238-4
Were you forwarded this newsletter?
LOVE IT.
If you liked what you read, consider signing up for your own subscription here:
