Omic.ly Weekly 27
June 2, 2024
Hey There!
Thanks for spending part of your Sunday with Omic.ly!
This week's headlines include:
1) Macrophages in lung cancer get their chance in the spatial omics spotlight
2) The microbiome and metagenome, what's the difference?
3) The ends of our chromosomes are a paradox
What you missed in this week's Premium Edition:
HOT TAKE: Grail suffers another setback as the UK NHS decides to wait until 2026 to move forward with a wider Galleri roll-out
Or if you already have a premium subscription:
The tumor microenvironment is a complicated place full of contradictions.
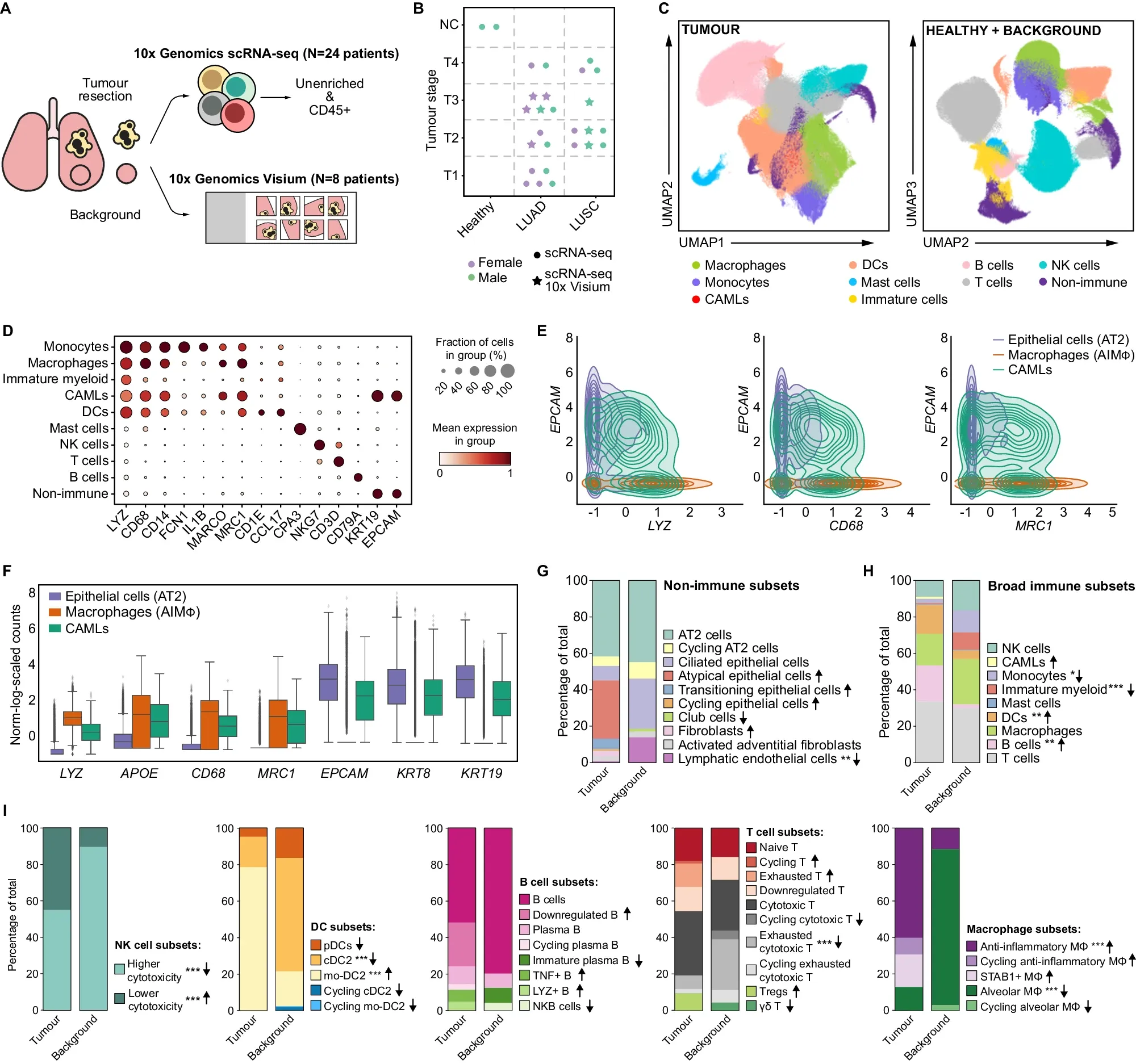
A new single-cell study on non-small cell lung cancer is helping us understand why that is.
Lung cancer is the second most common cancer globally and the leading cause of cancer death.
Of the two types of lung cancer, non-small-cell lung cancer (NSCLC) is the most prevalent type, accounting for about 85% of all lung cancer cases.
Small-cell lung cancer makes up the other 15%.
Both major types of lung cancer are associated with smoking or exposure of the lungs to agents that cause DNA damage, although there are genetic risk factors that can make individuals more susceptible.
NSCLC includes several subtypes, but adenocarcinoma (LUAD) and squamous-cell carcinoma (LUSC) are the two that are seen most often.
But NSCLC isn't just a mass of cells growing uncontrollably, it's a complicated cellular environment that we're now just beginning to understand.
This tumor microenvironment (TME) has been the focus of cancer research for many years now and we’ve learned a lot about the roles of various immune cells (like T cells and B cells), in patient prognosis.
We’re also now appreciating the role of macrophages (eat bacteria and other bad things) in lung cancer progression.
Tumor-associated macrophages (TAMs) have been shown to perform paradoxical functions.
They can promote tumor growth by suppressing the immune response or they can inhibit it by promoting inflammation and engaging in cytotoxic activity against cancer cells.
To better understand the role of these macrophages in NSCLC, the researchers behind today’s paper combined single-cell RNA sequencing (scRNA-seq) data from 25 patients with LUAD or LUSC with spatial transcriptomics from eight patients.
This combination allowed them to compare differences between tumor subtypes and also normal tissue.
What they found can be seen in the figure above: a) overview of the studies performed, b) umap comparing immune cell types found in normal and healthy tissue d-f) gene expression profiling of tumor associated immune cells g-i) further characterization of the state of the immune cells associated with healthy and normal tissues
This comprehensive TME profiling revealed important differences between the two subtypes of NSCLC.
The researchers found that each expresses different immune checkpoint proteins (inhibit the immune system’s ability to attack cancer) which means that they will likely respond differently to therapy.
This study provides a comprehensive dataset that identifies important molecular changes in the macrophage population within the tumor microenvironment, and could help guide therapeutic strategies against NSCLC.
###
De Zuani M, et al. 2024. Single-cell and spatial transcriptomics analysis of non-small cell lung cancer. Nat Commun. DOI: 10.1038/s41467-024-48700-8
The ‘Microbiome’ and the ‘Metagenome:’ what’s the $%# difference? Depends who you ask.

One of the things we come across during the development of new fields of science is that sometimes the vocabulary we have to describe the world isn’t sufficient.
When that happens, we need to invent new words (or mash up old words to make new words)!
But because this is science, it means we also spend a lot of time disagreeing on the definitions of words until we reach a consensus on what those words actually mean.
This usually happens once we have enough data to clearly define what it is that we are seeing!
But with respect to the ‘microbiome’ and the ‘metagenome,’ things are still a little murky.
There are two commonly used definitions for the microbiome, one has its roots in genetics and the other in ecology.
The genetics one is a mashup of microbi and ome (still pronounced micro-biome) and it is defined as all of the genomes of microorganisms found in a particular habitat.
The ecology one is a combination of micro and biome, and a biome in ecology describes all of the biotic and abiotic (environmental) factors that are found in a distinct geographic region.
So, a ‘microbiome’ in that version of the definition is just a micro version of a traditional biome.
It’s pretty confusing to have two very different definitions of a word.
But wait!
There’s more, because I haven’t told you about the metagenome yet!
Some people define it identically to the microbiome - it's the genomes of all of the microorganisms found in a particular habitat.
Others define it as all of the genetic information found in a habitat (inclusive of the host and any other environmental DNA from non-microorganisms).
Some say the microbiome is a subset of the metagenome.
Others claim that since the metagenome is just about genetics, it’s actually a subset of the microbiome!
Are you confused yet, because I certainly am!
But don’t worry, even microbiologists find this confusing, and in 2020 a controversial paper was published in the journal ‘Microbiome’ which tried to reconcile the competing definitions of all of these words.
In it the authors define the microbiome as all of the microbiota in a habitat inclusive of the theater of activity, meaning, they combined the two 'microbiome' definitions together.
In the case of microbiomes that occur on or in animals (like humans), this definition lumps them into the ‘theater of activity,’ too.
Which also means this definition eats all definitions of the metagenome because that definition can only be inclusive of 'genomes!'
While the definitions proposed by this paper provide order and logic to these words, we’re still probably decades from consensus on this!
###
Berg G, et al. 2020. Microbiome definition re-visited: old concepts and new challenges. Microbiome. DOI: 10.1186/s40168-020-00875-0
What if I told you that the ends of our chromosomes are a paradox and we didn't fully understand how human DNA was copied until 1989?
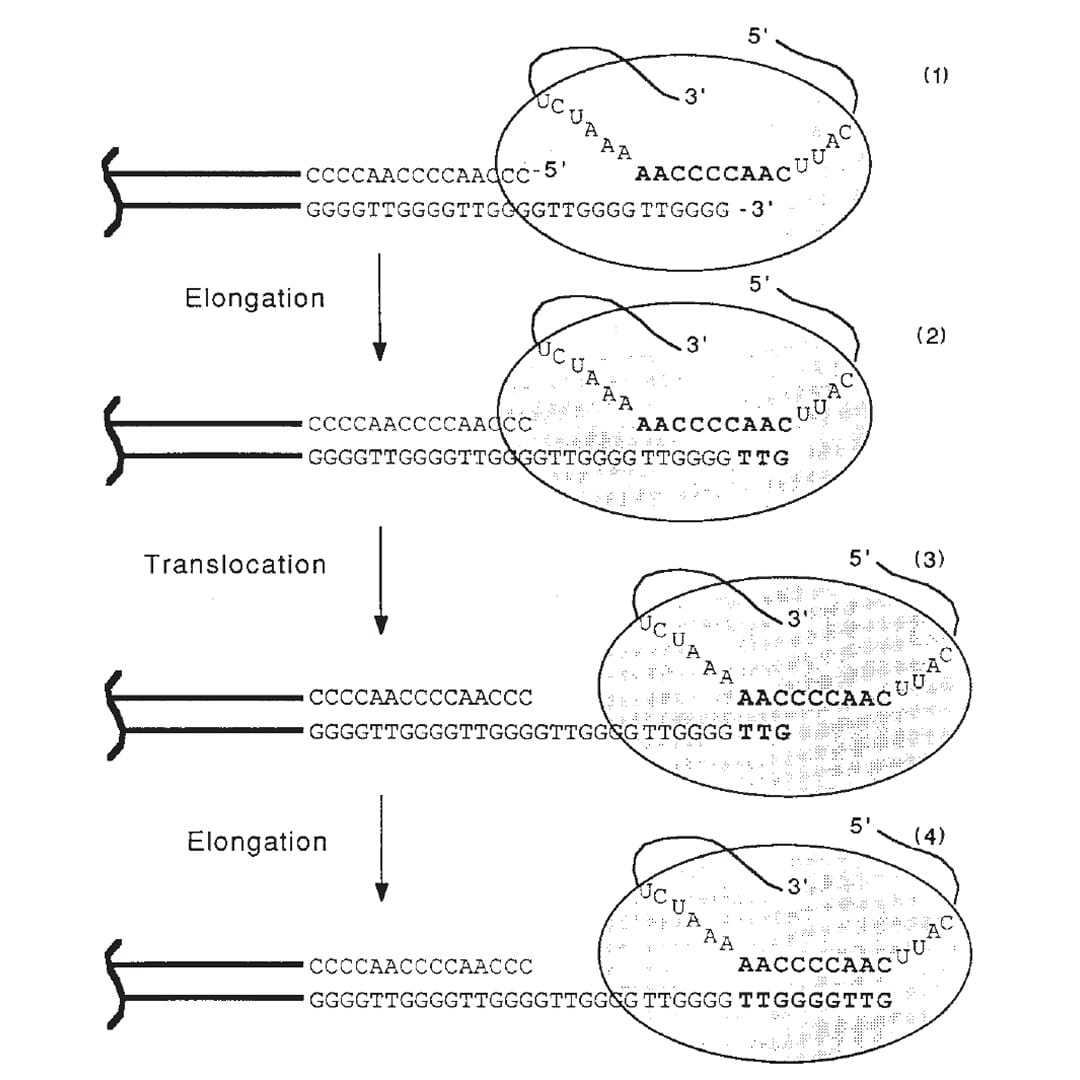
Meselson and Stahl showed in 1958 that replication of the DNA double helix, the process that copies DNA, is semi-conservative.
This means that the daughter double helix contains one newly synthesized strand and one old strand that serves as the template for copying.
During the process of replication, the parent DNA helix is unwound at an 'origin of replication' and the copying process begins.
Unlike humans, and other organisms that have a linear genome, bacteria don't have to figure out how to copy the ends of their genomes!
Bacteria have a circular genome so the process finishes where it began and ends with the creation of two circles.
But, if a genome is linear, there's a big problem when the machinery hits the end of a chromosome.
You see, replication only occurs in a single direction, 5'->3'. There's a 'leading strand,' which is made contiguously as the DNA is copied, and a 'lagging strand,' which is made by creating short fragments that are connected together using an enzyme called DNA ligase.
The problem is that once the leading strand gets to the end of a chromosome, the machinery that simultaneously creates the lagging strand doesn't have enough DNA at the end to sit on to fill in that last little fragment!
The lagging strand should always end up being just a little bit shorter than the leading strand, and so, a genome should get shorter every time it's copied!
Except it doesn't.
And that's due to a repetitive sequence at the end of our chromosomes called the telomere!
But we didn't really know what the telomere did until 1975 when Elizabeth Blackburn discovered that a highly repetitive nucleotide sequence (C-C-C-C-A-A) made up the telomeres of tetrahymena (a freshwater microorganism).
Blackburn had an inkling that this repetitive sequence was important and for the next 14 years she, along with help from Jack Szostak and Carole Greider, figured out how the ends of chromosomes are copied and preserved.
The figure above is the culmination of those years of experiments.
1) Shows the end of a chromosome and the telomerase enzyme (oval) with its RNA template bound to the TTGGGG of the telomere. Telomerase is a reverse transcriptase - these use RNA as a template to create DNA.
2) Elongation occurs, adding a TTG to the telomere.
3) Telomerase shifts down the strand (translocates) 6 bases to the new TTG.
4) Elongation occurs again.
This process of elongation and translocation is repeated multiple times, adding back sequence and preventing the shortening of the genome during replication!
Blackburn, Greider and Szostak were awarded the Nobel Prize for this discovery in 2009.
###
Greider CW, Blackburn EH. 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. DOI:10.1038/337331a0
Were you forwarded this newsletter?
LOVE IT.
If you liked what you read, consider signing up for your own subscription here:
