Omic.ly Weekly 26
May 26, 2024
Hey There!
Thanks for spending part of your Sunday with Omic.ly!
This week's headlines include:
1) APOE4: no longer just a risk factor for Alzheimer’s!
2) Bacteria are complicated. We invented pangenomes to better understand them.
3) We first discovered there was something funny going on with X-chromosomes in 1948.
What you missed in this week's Premium Edition:
HOT TAKE: Another summer of sequencing is upon us. Whose sparkle will beguile the hearts of suitors and dazzle the eyes of onlookers?
Or if you already have a premium subscription:
APOE4: no longer just a risk factor for Alzheimer’s, it's a genetically distinct form of the disease!
Alzheimer’s disease (AD) is the cause of ~ 70% of the cases of dementia in the US.
It's also the seventh leading cause of death.
The cause of AD is still largely unknown although there are genetic and environmental factors that contribute to the disease.
It is typified by the development of plaques in the brain that lead to the progressive loss of brain function.
The plaques are caused by the abnormal accumulation of the proteins amyloid beta (Aβ) and Tau.
Deficiencies in clearing these proteins are thought to result in inflammation of the brain that interferes with the connections between neurons and, ultimately, their death.
The best evidence we have for this mechanism is that the gene that leads to the creation of Aβ, Amyloid Precursor Protein (APP), is found on chromosome 21 and people with Down Syndrome (three copies of 21) generally show symptoms of AD by the age of 40.
This is further supported by Autosomal Dominant AD which is caused by mutations in APP and the proteolytic proteins PSEN1 and PSEN2.
These result in the abnormal overproduction of Aβ.
But, greater than 90% of cases of AD are sporadic (no family history or obvious genetic cause), although there are a number of genes that are risk factors for the disease.
One of those is Apolipoprotein E (APOE) which is involved in transporting lipids.
Genetic studies have identified 3 variants of APOE in humans: ε2, ε3, and ε4.
APOEε4 (or just APOE4) is associated with problems in regulating lipid metabolism and is often seen in people with hypercholesterolemia (too much cholesterol) and hyperlipidemia (too much fat in the blood).
It has also been associated with up to 80% of sporadic cases of AD and people who carry two copies of APOE4 have a 60% lifetime risk of developing disease.
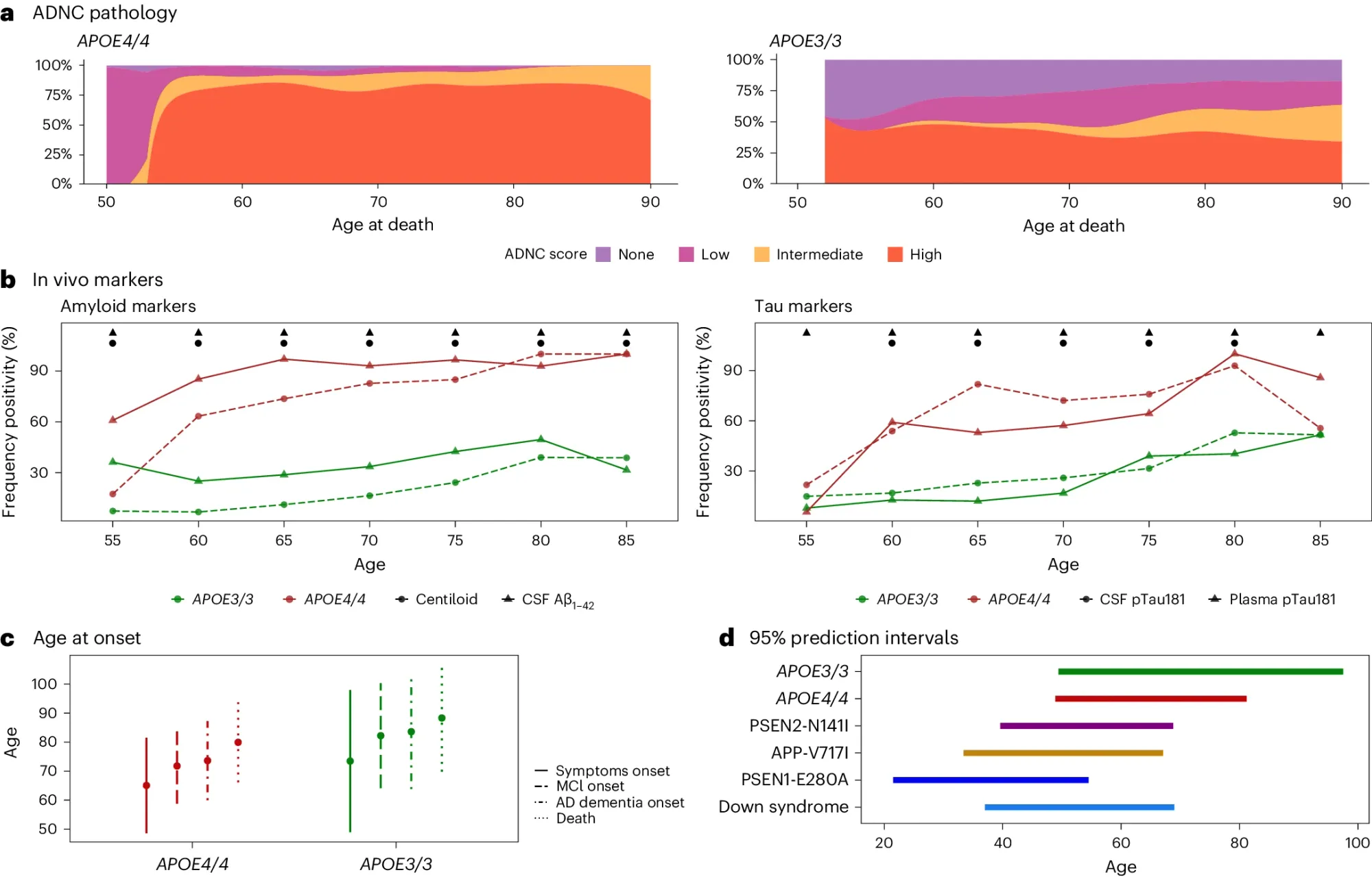
While a lot of questions still remain about how APOE4 can cause AD, the author’s of today’s paper make a very compelling argument that APOE4 represents a genetically distinct form of AD showing “near-full penetrance, symptom onset predictability and a predictable sequence of biomarker and clinical changes” that lead to disease.
They looked at the brains of >3,000 deceased AD patients and the clinical presentation of >10,000 individuals living with AD.
What they found (see the figure above) is that APOE4/4 AD patients a) have intermediate to high levels of neuropathology, b) >90% show biological hallmarks of the disease by age 65, c and d) have earlier disease onset, comparable to the other genetically distinct forms of AD.
This genetic distinction is important because it allows us to better inform patients of their risk, and gives us new tools to predict onset and track disease progression.
###
Fortea J, et al. 2024. APOE4 homozygozity represents a distinct genetic form of AD. Nature Medicine. DOI: 10.1038/s41591-024-02931-w
Pangenomes: the coolest new thing in human genetics got its start in bacteria. No, seriously!

If you're not quite sure what a pangenome is, don't worry, it's pretty simple!
It's all of the genes that are found in the genomes of each ‘clade’ of a species.
And clade is just an evolutionary term that identifies genetically distinct populations of individuals.
In humans, those are genetic ancestries but in bacteria we're talking about strains.
For example, when you hear about an E. coli ‘outbreak,’ what usually gets left out of the headlines is that it’s an outbreak of a pathogenic strain of E. coli.
You should know by now that ALL E. coli aren’t bad guys.
In the case of one of these food poisoning outbreaks, we’re usually talking about the O157: H7 strain of E. coli.
But there are other bacteria with strains that share similar opposing personalities.
One of those is Streptococcus agalactiae (or, group B strep) and can be found commonly in the vaginas of healthy women.
However, it can cause significant problems, or even infant mortality during pregnancy and the idea of a pangenome was first proposed in a study of the pathogenic strains of S. agalactiae!
With the falling sequencing costs that came with the invention of massively parallel sequencing, researchers were finally able to obtain full genomes from multiple strains of bacteria and compare them.
What we’ve found is that species of bacteria contain a ‘core’ of essential genes but that the genetic content at the strain level can vary significantly which produces the differences observed in strain level pathogenicity!
We’ve since sequenced a bunch more bacteria and expanded on this concept of a ‘core’ set of genes (found in >95% of genomes) to also include:
Shell genes - Found in 10-95% of genomes, these are genes are also typically shared with more than one strain
Cloud genes - Found in < 10% of genomes, these are strain specific genes
All of these genetic detours can be visualized as a ‘genomic graph’ showing an overlay of each genome and how they differ from one another.
So, why’s it important to characterize all of the genetic variation found in a species?
Because better understanding the strain level genetics can help explain why these bacteria can behave differently in different environments.
But things get extra weird in bacteria because not only do they have their own genome, but they also can have plasmids (extra DNA that can be shared).
They can also be infected by ‘bacteriophages’ (DNA viruses) which can change the genetics AND the behavior of these bacteria!
That’s in addition to the mutations that occur to their genomes as they grow and replicate.
Things get only moderately less complicated as we transition the concept of the pangenome to other organisms like humans.
We may not have plasmids that can do goofy things with our genetics, but viruses and environmental pressures have helped shape our genetic ancestry in ways we’re only just beginning to appreciate!
Biologically female mammals have two X chromosomes, but what might surprise you is that one of those X's is turned off.
The first person to recognize something different about female animal cells was Murray Barr.
In 1948, he discovered in cats that female cells have a 'nucleolar satellite' which appeared as a black dot in the nucleus.
This was odd because a similar structure could not be found in males.
It was hypothesized that this 'satellite' material was related to sex chromosomes and it later became known as the 'Barr Body.'
Over the span of 11 years, the Barr Body was used to easily discriminate the biological gender of cells.
But in 1959, Susumu Ohno showed that the Barr body was actually a condensed X chromosome!
Two years later, Lyonization, or X-inactivation, was first proposed by Mary Lyon after she observed a correlation between which X ended up being condensed and the coat color of female mice.
Ok, that’s great, but why’s it important for one of the X chromosomes to be turned off?
Dosage.
This is a fundamental concept in genetics and gene duplications or deletions can increase or decrease how many copies of each gene are in the genome.
Issues with dosage are most apparent in diseases like Down Syndrome where individuals have an extra copy of chromosome 21.
This imbalance results in the production of more protein than is required for normal cellular function and also is responsible for the observed phenotypes.
So, X-inactivation ensures that just the right amount of protein is produced from the code written in the X chromosomes!
While genetics is pretty good at correlating genetic changes with broader phenotypes, it can’t prove a direct relationship.
And so the molecular sleuthing began in 1991 with Huntington Willard and his team who located the 'X inactivation center' and discovered a long non-coding RNA (lncRNA) that seemed to encapsulate and condense the inactived X chromosome.
They named this lncRNA the X-inactive specific transcript or, XIST, for short.
But there was still a nagging question about whether this actually turned off gene expression.
A good chunk of that answer came as a result of the work of Jeannie Lee, who, in 1996, was a postdoc in Rudolf Jaenisch’s lab.
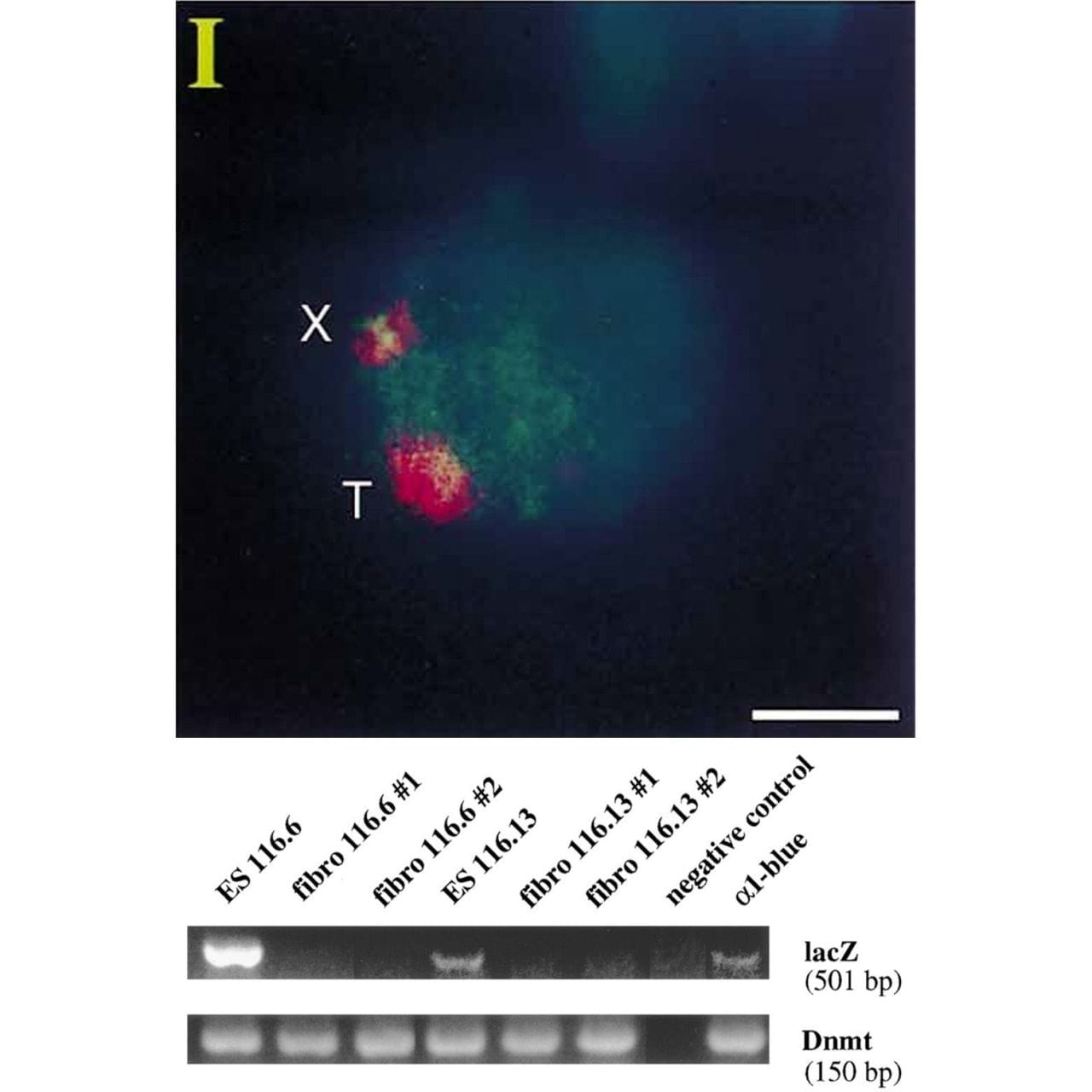
Lee copied the XIST sequence into a yeast artificial chromosome (YAC) that expressed a common ‘reporter’ gene, LacZ.
When introduced into mice, this YAC (T) and the mouse X chromosome (X) both expressed XIST RNA (red cloud).
But the proof that XIST represses gene expression came when looking at LacZ.
The gel shows that LacZ is expressed in embryonic stem cells (Lanes 1 and 4) but not in differentiated fibroblasts (Lanes 2,3,5 and 6)!
This work represents the first direct evidence of a functional role for XIST and its broader involvement in repressing gene expression from the inactived X chromosome.
###
Lee JT, et al. 1996. A 450kb Transgene Displays Properties of the Mammalian X-Inactivation Center. Cell. DOI: 10.1016/s0092-8674(00)80079-3
Were you forwarded this newsletter?
LOVE IT.
If you liked what you read, consider signing up for your own subscription here:
