Omic.ly Weekly 25
May 19, 2024
Hey There!
Thanks for spending part of your Sunday with Omic.ly!
This week's headlines include:
1) Are personalized vaccines the next big thing in cancer therapy?
2) Genetic Determinism: What it is and how we can avoid it with multi-omics
3) Insulin: It was believed to exist, but everyone who tried to isolate it failed
What you missed in this week's Premium Edition:
HOT TAKE: AlphaFold, the second coming of Christ or just another useful tool in the life sciences?
Or if you already have a Premium subscription:
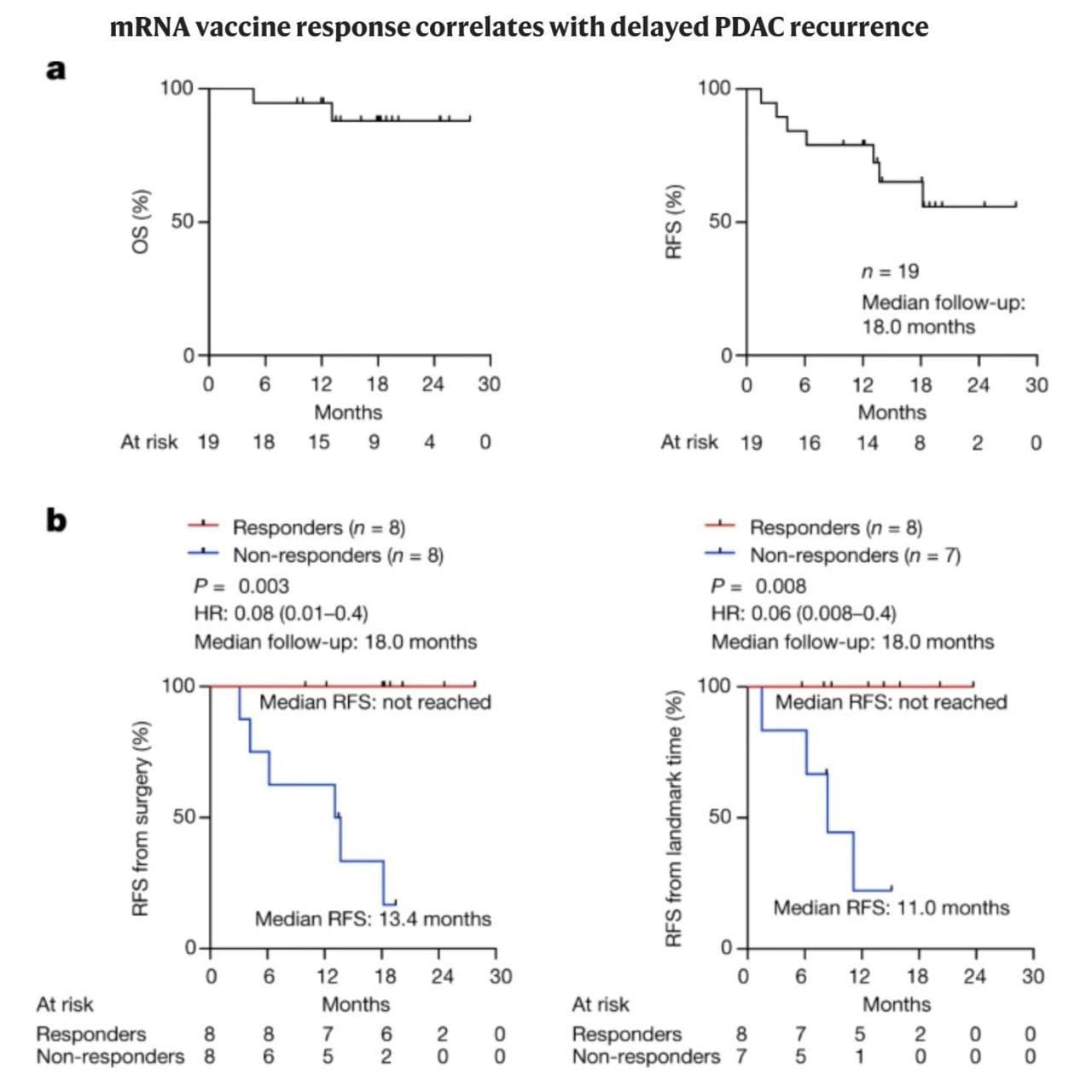
Are personalized vaccines the next big thing in cancer therapy? The figure below kind of speaks for itself.
The therapeutic potential for mRNA technology goes well beyond its use in vaccines for infectious diseases!
Because mRNA is easy to program, it can be used for a variety of other personalized therapies.
One of the most intriguing of these is to use it to eradicate cancers.
This is possible because cancer cells are pretty messed up and they make a lot of weird proteins that can be recognized by the immune system!
These are often referred to as ‘neoantigens.'
That word might look intimidating.
But don't worry, neo is just ‘New’ and antigen is ‘something that can be recognized by the immune system.’
So, how can we use these ‘neoantigens’ to coax the immune system to destroy cancer?
We can make mRNAs that express them, stick them in a vaccine, and prime the immune system to then specifically recognize cancer cells!
But wait, if cancer cells already express these neoantigens, why doesn’t the immune system destroy cancers without the priming?
Mostly it’s because tumors start out small and don’t activate enough cells to mount a strong immune response.
As tumor cells grow, the immune system sees these same antigens over and over again so it begins to tolerate them.
The purpose of a cancer vaccine is to supercharge the immune system to recognize tumors as invaders!
The author’s of today’s paper did just that and targeted a notoriously hard cancer to beat:
Pancreatic ductal adenocarcinoma or PDAC.
This cancer is lethal in 88% of patients and 90% of PDACs recur (come back) 7-9 months after surgery.
So, the need for an alternative therapy is significant.
In this study, patients with a PDAC that could be surgically removed were recruited and then treated with a combination of an immune checkpoint inhibitor, an individualized mRNA neoantigen vaccine, and a chemotherapy regimen.
The mRNA vaccine was designed against tumor-specific neoantigens that were discovered through whole exome and RNA sequencing of the removed PDAC tissue.
Ultimately, 19 patients were administered the checkpoint inhibitor, 16 went on to receive a personalized vaccine, and 15 were given the chemotherapy regimen.
The overall survival and relapse free survival can be seen above in (A), which in itself is a significant improvement over the current standard of care.
But when broken out by vaccine responders and non-responders (B), the results are nothing short of remarkable.
While only 50% of vaccine recipients responded, they were 100% relapse free beyond 18-months.
Despite the small sample size of this initial study, the future of personalized cancer vaccines seems bright.
And, with improved methods for neoantigen identification, hopefully we can get that responder rate closer to 100% too!
###
Rojas LA et al. 2023. Personalized RNA neoantigen vaccines stimulate T cells in pancreatic cancer. Nature. DOI:10.1038/s41586-023-06063-y
One thing we constantly battle in genomics is, “What does it all mean?”

As with everything, it's complicated.
We have 5 million variants in each of our genomes with about 100,000 of those falling in the coding regions of genes.
Of those, a couple hundred are damaging but the majority are what we refer to as variants of unknown significance (VUS).
The process of determining whether those variants cause disease (are pathogenic) or benign (don't cause disease) isn’t incredibly sophisticated.
It’s generally done by looking at families with a history of disease, finding what variants are common in those affected and then deciding with some degree of certainty based on the function of a gene whether or not a variant fits well with the symptoms.
One of the most used databases for assessing whether a variant might cause a disease is ClinVar, and underpinning the sophistication of this process, ClinVar ranks the certainty of a classification with gold stars.
Yes, your kindergarten teacher was onto something after all.
So, it’s probably fair to say we’re still pretty early in the game of figuring out all of this stuff, but too often in genomics we try to find signal in the noise.
Because if there’s a variant, it has to DO something!
And unfortunately, this means we stumble into the pernicious problem of genetic determinism or this idea that the code written in your genome is the only thing that determines whether you will develop a disease.
We realized a while ago that genetics really is only a component here and other genetic, environmental and epigenetic factors all contribute to whether a disease develops (this is called penetrance) and similarly, these factors can also determine the severity of disease in an individual (this is sometimes referred to as expressivity).
However, historically, these measurements were done in families who were affected by a genetic disease so the estimations of penetrance have always been assumed to be over estimates.
Today we can do better because we now have huge population level datasets.
Recent studies have found that about 8% of people carry a pathogenic mutation, but only 7% of those were symptomatic in the EHR.
However, the penetrance of individual mutations varies greatly, for example, on average, 38% of BRCA mutation carriers develop breast cancer.
These results highlight the importance of approaching disease risk associations carefully, especially when reporting results in healthy people.
The genome only ever represents what 'could be,' but observing changes in our other 'omes,' which are the functional product of the genome, can help us better predict when genetic variations might actually manifest as disease.
Because genetics isn't deterministic and for us to truly get a handle on disease and disease progression, we need to marry what we see in the genome with what we can measure in the other 'omes.'
Insulin: It was believed to exist, but everyone who tried to isolate it failed.
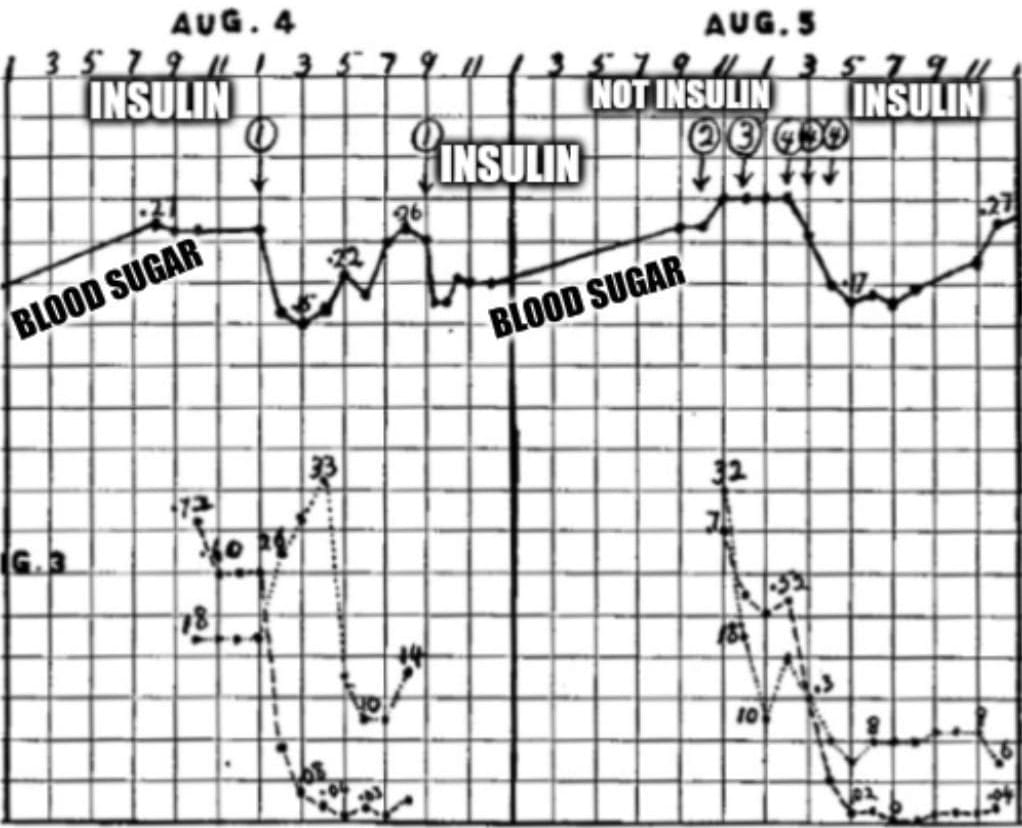
It was finally captured in 1921.
Many decades worth of medical observation and dissection showed that problems with the pancreas caused diabetes.
The classical symptoms are high blood sugar, glycosuria (excretion of sugar in the urine), ketosis (burning fat instead of sugar), seizures, and, if untreated, death.
It was discovered that animals who suffer from diabetes naturally lose their pancreatic islet cells, and in 1909, Jean de Meyer, and in 1913, Edward Sharpey–Schäfer, suggested that a hormone was secreted from the islet cells to control glucose metabolism and its uptake from the bloodstream.
Sharpey–Schäfer named this substance 'insuline.'
But, this is not the only function of the pancreas.
It also produces trypsin, an enzyme that helps digest proteins in the stomach.
This made things complicated because everyone who tried to isolate insulin ultimately failed because it was immediately chopped up during extraction by trypsin!
Fortunately, Frederick Banting, a Canadian surgeon, came across an article describing how the blockage of the pancreatic ducts in a patient resulted in the death of the acinar cells (the ones that pump out trypsin), but preserved the islets with no sign of the development of diabetes.
Banting, who had no formal scientific training, thought that a good way to isolate insulin might be to 'ligate' or tie off the pancreatic ducts to kill all the acinar cells and then grind up what was left over.
He pitched his grand idea to JJR Macleod, a professor of physiology at the University of Toronto, who had a lab and access to research animals.
Macleod was skeptical, but he was also going on vacation, so he gave Banting the keys to the lab, 10 dogs, and a coin flip determined that Charles Best would help Banting.
Best was also tasked with making sure the lab didn't burn down while Macleod was away.
7 dead dogs later, Banting and Best had perfected their technique, and generated the data that can be seen above.
What's displayed is two days worth of blood glucose readings (solid black line) from a dog whose pancreas was removed (causes diabetes).
The numbered circles are injections of ground up organ extracts: 1. Refrigerated ligated pancreas, 2. Liver, 3. Spleen, and 4. Fresh ligated pancreas.
Here you can see that ligated pancreatic extracts (1 and 4) cause a sharp reduction in blood glucose.
Insulin had finally been isolated!
Further work by James Collip revealed that highly purified insulin could be made from chilled pancreas (no ligation) and extraction in 90% ethanol.
This process was optimized by Eli Lilly for use on pig and cow pancreas which produced the majority of insulin until a synthetic was developed 56 years later in 1978.
###
Banting FG, Best CH. 1922. The internal secretion of the pancreas. J Lab Clin Med. DOI: 10.1111/j.1753-4887.1987.tb07442.x
Were you forwarded this newsletter?
LOVE IT.
If you liked what you read, consider signing up for your own subscription here:
