Omic.ly Weekly 24
May 12, 2024
Hey There!
Thanks for spending part of your Sunday with Omic.ly!
This week's headlines include:
1) Ever get lost in your own heart? Have no fear, there's a new atlas for that!
2) The gut-brain axis: now with microbes
3) Nettie Stevens discovered sex chromosomes in 1905
What you missed in this week's Premium Edition:
HOT TAKE: Spatial omics may give that dying NovaSeq of yours a second life!
Already have a premium subscription?
That beating sensation you feel in your chest is the product of billions of cells working together to pump blood throughout your body.
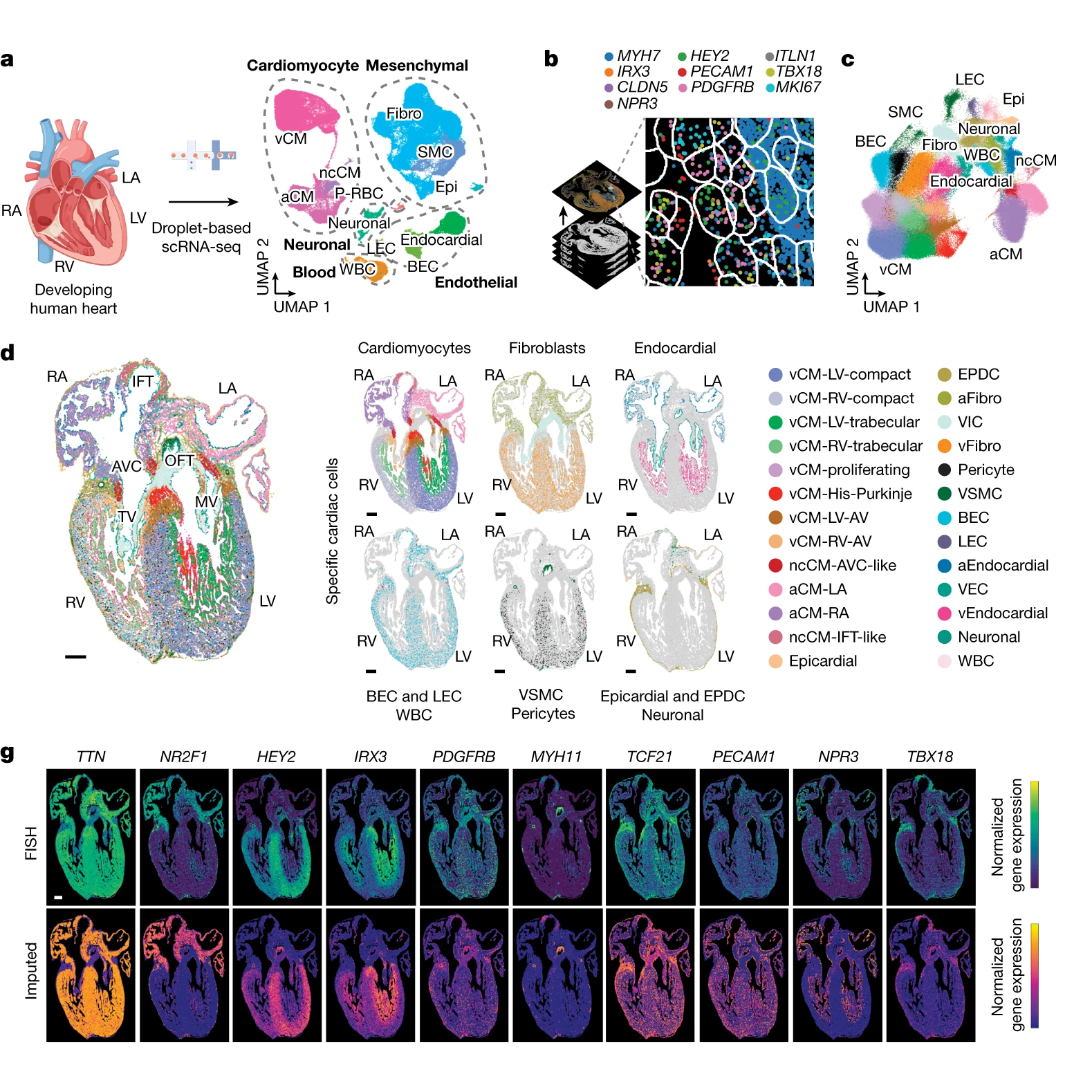
We now have a better idea of how those cells accomplish this!
You might not think about this very often, but your heart is critical for keeping ALL of the cells in your body alive.
It’s probably the most important organ involved in bringing nutrients to cells and removing waste from the cellular environment.
That’s not to discount the lungs, gut, liver and kidneys!
But without our blood pump, none of those organs would be able to do their jobs either.
The heart is also an incredibly complex organ composed of various structures that are essential for its rhythmic beating.
Disruption of these structures during development can lead to congenital heart disease or structural heart diseases like hypertrophic cardiomyopathies and valvulopathies later in life.
But, understanding how these diseases develop has been tough.
This is because the heart has a ton of cells that all perform slightly different functions!
These include cardiomyocytes (muscle), cardiac fibroblasts (structural support), endothelial cells (cover the inner and outer surfaces), smooth muscle cells, neuronal cells, and immune cells.
But determining the composition and interactions of these cells and how they form and maintain the crucial structures and functions of the heart has remained a challenge.
Fortunately, we now have new single-cell techniques to tease apart the answers to these questions!
The researchers behind this week’s paper combined single-cell RNA sequencing (scRNA-seq) and multiplexed error-robust fluorescence in situ hybridization (MERFISH) on developing human hearts.
The figure above shows the major findings of the paper including an overview of the a) scRNA-seq and b and c) MERFISH techniques along with how each clustered cells through gene expression mapping.
In d) MERFISH gene expression maps were overlaid visually to show where all of the major classes of heart cells are located spatially.
And g) validates the performance of MERFISH and scRNA-seq by comparing the gene expression levels observed by each technique.
Overall, cardiomyocytes and non-cardiomyocytes showed regional and structural diversity, with distinct populations occupying specific anatomical domains.
Further analysis revealed the formation of cellular communities within the heart, where specific combinations of these cells are organized to perform the crucial functions of each heart region.
Together this dataset provides a high-resolution single-cell molecular and spatial cardiac cell atlas.
It also helps to explain how the distinct cell types of our hearts organize into the cardiac structures essential for the rhythmic ‘lub-dub’ that keeps us all alive!
###
Farah EN, et al. 2024. Spatially organized cellular communities form the developing human heart. Nature. DOI: 10.1038/s41586-024-07171-z
We’ve known since the time of Pavlov that there’s a connection between our gut and brain; now we know our microbiomes play a role in that game of telephone too!

If you’re not familiar with Pavlov, he won the Nobel in 1940 for discovering that the smell or sight of food triggers the stomach to secrete all the goodies that are responsible for breaking down the food we eat.
This was the first clear link that our brains and our gut communicated.
But it’s not just a one-way conversation!
Our guts also communicate back to the brain to tell us when we’re full or hungry.
More recently we’ve come to realize that the microbes that live in our gut also participate in these conversations in complex ways.
Our gut harbors the largest of our microbiomes and contains trillions of bacteria, fungi, viruses, and archaea.
Evidence suggests that the composition of this microbiome can affect not only how we digest food but potentially our moods, behavior, or the development of neurological disease!
The gut microbiome can do this through interactions with the gut-brain axis which includes:
Autonomic Nervous System: Controls bodily functions such as breathing and digestion, basically all of our bodily functions that happen without us ‘thinking’ about them.
Enteric Nervous System: Refers to the nerves of the gut that function in digestion and ‘moving’ food and waste through it.
Neuroendocrine System: Uses hormones as signaling molecules to control bodily functions but with respect to the gut these discussions are focused primarily on stress responses through the Hypothalamic-pituitary-adrenal axis.
Immune System: Determines how we respond to and clear infections.
Metabolism: Controls how and what we’re able to digest to produce energy or build the precursors that make up all of the molecules in our cells.
The microbes in our gut can communicate directly through all of these pathways because they produce signaling molecules like neurotransmitters, hormones, immune molecules (chemokines and cytokines), amino acids, and microbial metabolites.
These in turn can influence neuronal cells, the activities of our neuroendocrine system, host immune responses, or metabolism.
They can also travel through our bloodstream to interact with the brain directly!
Recent research has also highlighted the importance of these communication pathways in neurological conditions such as anxiety, autism spectrum disorder, and depression.
Unfortunately, we’re still in the early stages of deciphering cause and effect here!
And important questions about whether certain microbes cause neurological disease, or if individuals with neurological disease establish different microbiomes after onset, still need to be answered.
However, it’s clear that our microbiome and the gut-brain axis are in constant communication.
Now we just need to figure out if we can manipulate those conversations to promote health or prevent disease!
Nettie Stevens, a former school teacher turned geneticist, discovered sex chromosomes in 1905. Here's her story:
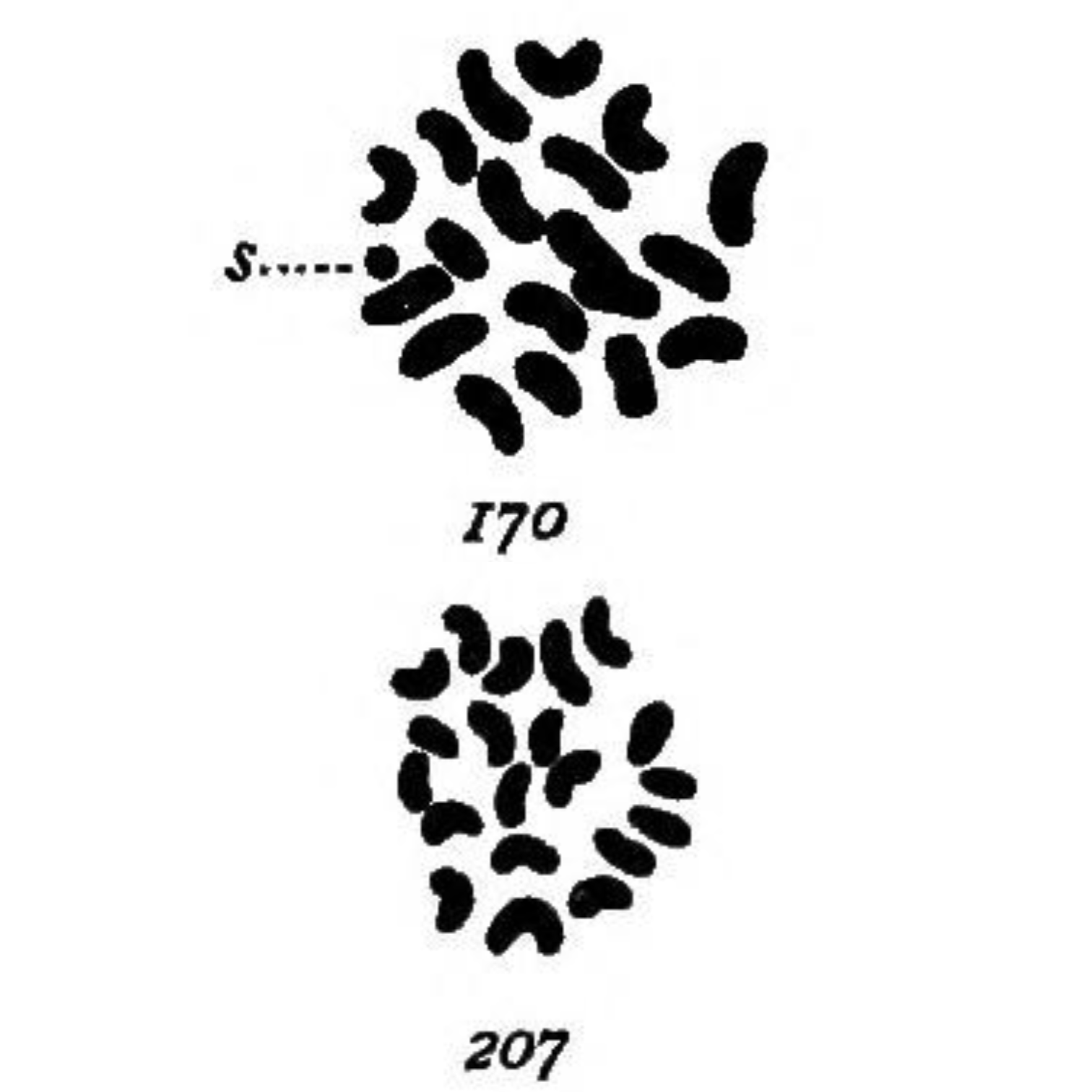
The late 1800s were a turning point in American history for women.
The end of the Civil War marked the start of the women's rights movement and granted them significantly more control over their own lives.
That being said, women were still expected to be teachers or home makers but they won more personal freedom including better access to education.
Stevens took advantage of these expanded liberties, and started her educational pursuits at age 10 studying to become a school teacher.
In 1883, she began a decade-long career in education, filling roles both as a teacher and as a librarian.
However, this wasn't her life's dream, and in 1896, at the age of 34, she had saved enough money to enroll at Stanford University earning both her bachelor's and master's degrees in 1900.
It was during her summer studies that she took a keen interest in cytology while working at the Stanford Marine Lab.
Here she spent her time glued to a microscope and published her first paper on the life-cycle of ciliates in 1901.
Stevens then left Stanford to continue her scientific studies at Bryn Mawr beginning her dissertation work under the guidance of Thomas Hunt Morgan.
You might be familiar with him.
Morgan received the Nobel Prize in 1933 for his work elucidating the role of chromosomes in heredity.
Interestingly, Morgan was initially skeptical of heredity, particularly as it related to sex determination.
At the time, there were two competing hypotheses:
One being that sex was determined by environmental factors, like temperature, and another that sex was inherited as a trait.
In spite of the lingering questions on this topic, Stevens successfully completed her dissertation in 1903, and received an award from the Carnegie Institution to study how sex is determined.
With this funding, Stevens meticulously detailed the cellular structures of the reproductive organs of multiple insects.
The most striking of these being her drawings in 1905 (see above) of the chromosomes found in mealworms (Tenebrio molitor) where she observed:
‘In both somatic and germ cells of the two sexes there is a difference not in the number of chromatin elements, but in the size of one, which is very small in the male (170-s) and of the same size as the other 19 in the female (207).’
Even though she was the first to make this discovery, Thomas Hunt Morgan and Edmund Beecher Wilson are often incorrectly given the credit.
Tragically, breast cancer cut her scientific career short at the age of 50.
But Nettie Stevens' contributions to our understanding of sex determination, along with her exquisite drawings, were the definition of groundbreaking in the field of genetics.
###
Stevens NM. 1905. Studies in Spermatogenesis with Especial Reference to the Accessory Chromosome. Carnegie Institution.
Were you forwarded this newsletter?
LOVE IT.
If you liked what you read, consider signing up for your own subscription here:
