Omic.ly Weekly 23
May 5, 2024
Hey There!
Thanks for spending part of your Sunday with Omic.ly!
This week's headlines include:
1) No DNA mutations needed. Epigenetic changes can cause cancer.
2) Pathobionts: The opportunists hiding in our microbiomes.
3) Barbara McClintock discovered a little thing called the transposable element in 1950
What you missed in this week's Premium Edition:
HOT TAKE: Did the FDA chicken out with its push to regulate lab developed tests? Kinda.
Or if you're already a Premium Subscriber:
No DNA mutations needed. Epigenetic changes can cause cancer. Woah!
Cancer is generally understood to be the result of dysregulation of cellular processes that lead to the uncontrolled growth of cells.
For the longest time, our focus on what causes cancer has been centered on DNA mutations that result in the activation of oncogenes (promote cancer) and the repression of tumor suppressors (don't promote cancer).
But in our study of oncogenes and tumor suppressors, we noticed that many of these genes were involved in epigenetic regulation of the genome.
Epigenetic changes are loosely defined as non-sequence alterations that regulate gene expression.
So, we’re talking about all of the genes involved in how the information in our DNA is accessed (or not accessed).
And these include proteins that methylate DNA, modify histones (the little hockey pucks that wrap up and organize our chromosomes), move around histones, or change how our chromosomes fold into larger 3D structures.
While it makes sense that epigenetic regulation of gene expression is a major component of the development of cancer, we have, until now, not shown that epigenetic modifications alone were sufficient to create tumors.
In today’s paper, the researchers focused on the activities of Polycomb group proteins (PcG) which make up the Polycomb repressive complex 1 and 2 (PRC1 and PRC2).
These are protein complexes that are involved in maintaining repressive (H2AK118Ub and H3K27me, respectively) marks on histones.
PcG proteins are highly conserved in eukaryotes and disruption of these complexes has been associated with developmental abnormalities and cancer.
But can disruption of PcG complexes and the loss of repressive marks on chromatin cause cancer?
The answer to that seems to be yes!
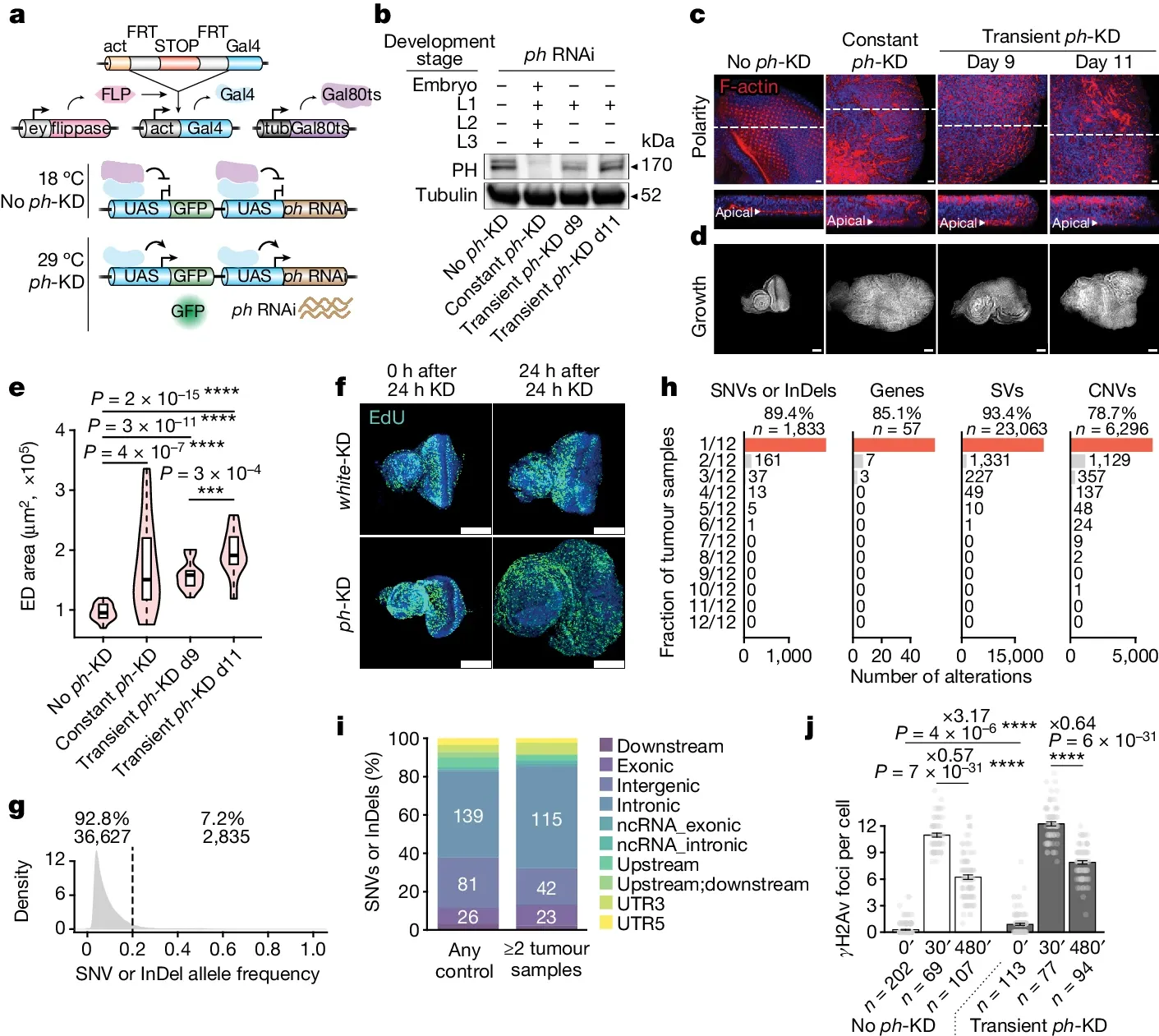
The figure below shows an experiment that was carried out in flies (share many of the same proteins as humans).
And because global loss of PRC complexes can cause bigger problems, they did these experiments in a very specific tissue, the eye disc (ED).
a) experimental setup where removal of ph (a component of PRC1) is controlled by exposure to different temperatures. b) the level of ph protein expression after the different temperature treatments. c and d) the development of ED tumors after treatment. e and f) short term loss of epigenetic control is enough to cause tumor development. g, h, and i) the tumors are likely NOT the result of DNA mutations. And j) DNA repair was not impacted.
Together these results indicate that it is possible for epigenetic changes to be solely responsible for the development of tumors.
These results also highlight the importance of epigenetic changes in cancer development.
And they underscore the utility of studying more than just DNA mutations in the genome.
###
Parreno V., et al. 2024. Transient loss of Polycomb components induces an epigenetic cancer fate. Nature. DOI: 10.1038/s41586-024-07328-w
The microbes we co-exist with can be friends, foes, or just along for the ride. They can also switch between being all of those things…

The external and internal surfaces of our bodies are covered by microbes.
Actually, most surfaces we interact with on a daily basis are covered in them!
And the viruses, fungi, bacteria and archaea that call us home help to make up our microbiomes.
The largest of these is the gut microbiome but we also have skin, oral, urogenital, and respiratory tract microbiomes.
Each of these is colonized by different combinations of microbes and a ‘microbiome’ describes those community members, their habitat, and how they interact with one another.
But we’re still in the early stages here and our understanding of our microbiomes is in its infancy.
That means that a lot of the terms that we use to describe microbiomes (including the word microbiome!) are still being hotly debated.
We also haven’t settled on a vocabulary because so many of the interactions that we see between microbes and their hosts are different from what we’ve observed in plants and animals!
While there are many shared common relationships within the microbiome, there are also new relationships that we’re still trying to understand and define.
We’ve certainly observed the traditional symbiotic relationships (parasitism, mutualism, commensalism, etc), but in microbiomes, microbes can blur the lines of what’s expected.
Some microbes operate as opportunists and can quickly switch from commensal (benefit but do no harm) to mutualistic (everyone benefits) to parasitic (they benefit while harming everyone else).
This highlights the importance of microbial ‘homeostasis’ or community stability because changing environmental conditions can allow these microbes to go from being friends to enemies!
But even that binary description isn’t really accurate and that’s a symptom of us not knowing enough about anything yet to properly define what’s going on here!
But that hasn’t stopped us from trying!
And we’ve recently begun referring to these opportunistic microbes as 'pathobionts.'
A few examples here include:
Bacteroides fragilis - Implicated in Irritable Bowel Syndrome and Colon Cancer
Fusobacterium nucleatum - Typically found in the oral cavity but can cause Gum Disease and has been associated with Colon Cancer
Enterococcus faecalis, Clostridioides difficile, and Klebsiella oxytoca - Have all been shown to be associated with the development of Colitis
Escherichia coli and Proteus mirabilis - Have been seen associated with Crohn’s disease
All of these bacteria are present in our microbiomes or we’re exposed to them consistently in our daily environments.
But how these pathobionts lead to health or disease is still an ongoing area of intense research.
One thing is clear though, and that’s that microbiomes are complicated!
While everyone else was distracted by the structure of DNA, Barbara McClintock was discovering a little thing called the transposable element.
The 1950's were a turning point in the fields of molecular biology and genetics.
For many years, scientists studied genetics but weren't entirely sure how it all worked on the molecular level.
They were aware of chromosomes, knew that they were composed of DNA and protein, but there was still a big question about the functional role of those two components in inheritance.
Fortunately for McClintock, who studied genetics in maize (corn), it didn't matter much what the genetic material was but that it could be tracked and its effects seen in the offspring of her crosses.
As an early cytogeneticist (scientist that studies chromosomes), McClintock developed many of the foundational techniques that were required for studying chromosomes in maize and she produced the first genetic maps of all 10 chromosomes.
Her early work was focused on studying inheritance and she was one of the first to observe and describe the phenomenon of homologous recombination, or crossing-over, which is when two chromosomes exchange genetic information.
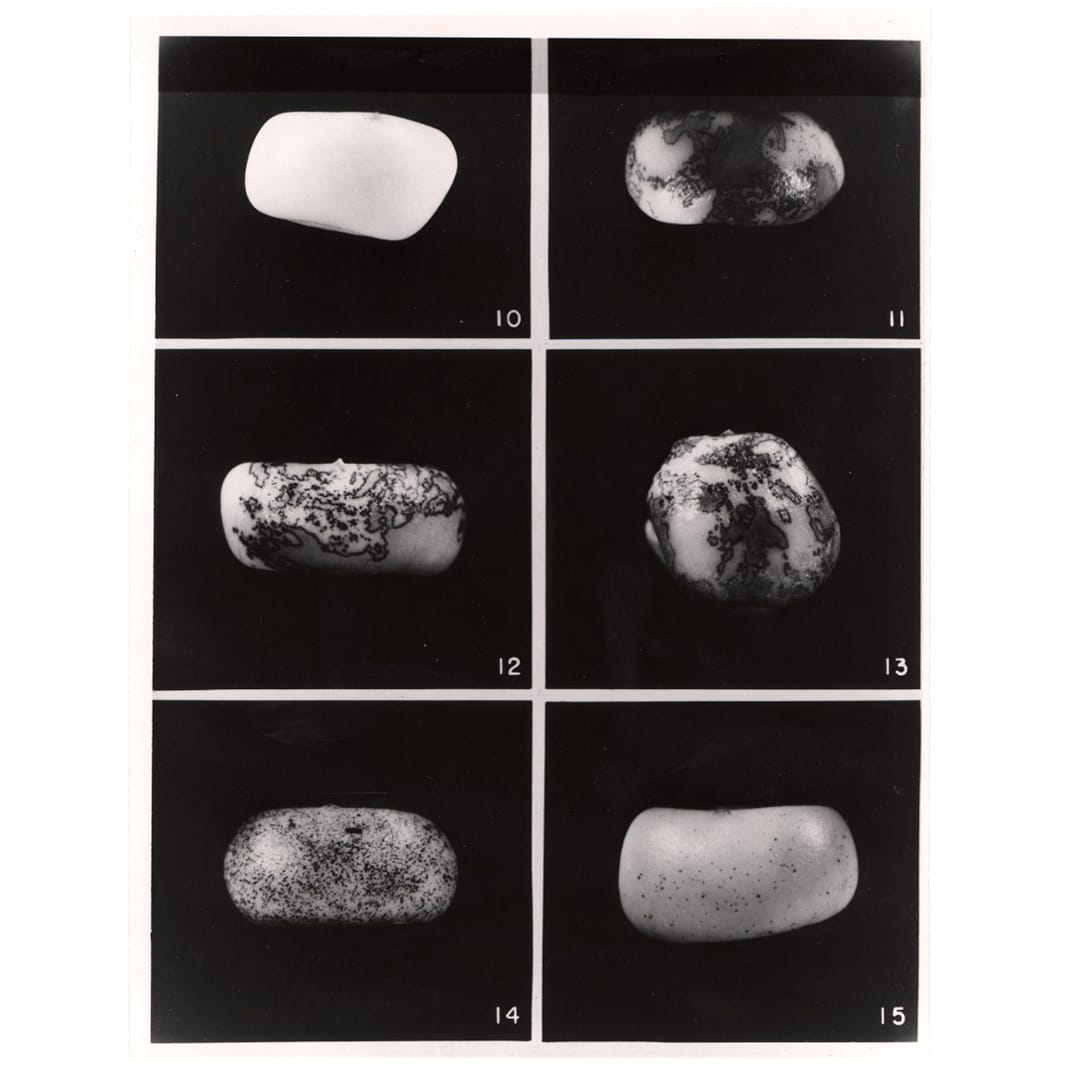
She published this work in 1931 and continued to study maize genetics until her curious observation that pieces of chromosome 9 appeared to move around the genome.
But these pieces, Ac (activator) and Ds (inhibits pigment production), didn't just move around, Ac seemed to control Ds, and ultimately the color patterns of the maize kernels!
The figure above displays the kernels that McClintock described in, 'The origin and behavior of mutable loci in maize.'
Kernel 10 - no Ac, 11-13 - 1 copy of Ac, 14 - 2 copies, 15 - 3 copies.
We know today that 66% of the human genome and 85% of the maize genome is made up of these mobile 'transposable' elements and they are key in our understanding of epigenetics, but back in 1950, this discovery was problematic.
At the time, it was thought that genomes were static and ordered; pieces of them did not move around.
McClintock's findings were so poorly received that she stopped formally publishing them after 1953 because she thought her colleagues just weren't interested.
Fortunately, her work was 'rediscovered,' or, more accurately, 'understood,' in the 1970's when similar elements were found in bacteria, flies and humans.
She was awarded the Nobel in 1983 for the discovery of transposable elements, which was 3 decades after her original description of them.
McClintock's story in science isn't dissimilar from that of other prominent women, like Rosalind Franklin.
However, unlike Franklin, McClintock lived long enough to finally be properly recognized for her historic achievements.
###
McClintock, B. 1950. The origin and behavior of mutable loci in maize. PNAS. DOI: 10.1073/pnas.36.6.344
Were you forwarded this newsletter?
LOVE IT.
If you liked what you read, consider signing up for your own subscription here:
