Omic.ly Weekly 22
April 28, 2024
Hey There!
Thanks for spending part of your Sunday with Omic.ly!
This week's headlines include:
1) The evolutionary history of coffee is probably more complicated than you think.
2) Clinical applications of the microbiome are still very much a work in progress.
3) The first human "Mendelian" disease was described in 1902.
What you missed in this week's Premium Edition:
HOT TAKE: We should all be a little worried about life insurance companies doing polygenic risk scoring on customers.
That dark and delicious brew billowing clouds of irresistible coffee incense next to you is most likely the product of Coffea arabica.
60% of the coffee beans that make it into our mugs are ‘Arabica.’
But Arabica itself is a hybrid of two other coffee species: C. canephora and C. eugenioides.
Things get weird here, though, because plant genomes do crazy things during hybridization.
In the case of Arabica, it becomes polyploid, which is the result of whole genome duplication.
Arabica’s parents have two sets of chromosomes and Arabica is the proud owner of 4 sets.
It’s allotetraploid!
Which just means that it’s made up of genomes from two different species.
And this divine combination is thought to have occurred hundreds of thousands of years ago in Ethiopia.
But it was first discovered in 850 CE by a goat herder in Yemen (hence the name) who noticed that whenever his little bleaters nibbled on Arabica’s berries they became exceptionally energetic.
According to the legend, he reported this to a local monk who turned the berries into a drink!
We now produce about 25 billion pounds of coffee every year which has made it one of the most popular beverages in the world.
It has also made it one of the most important crops!
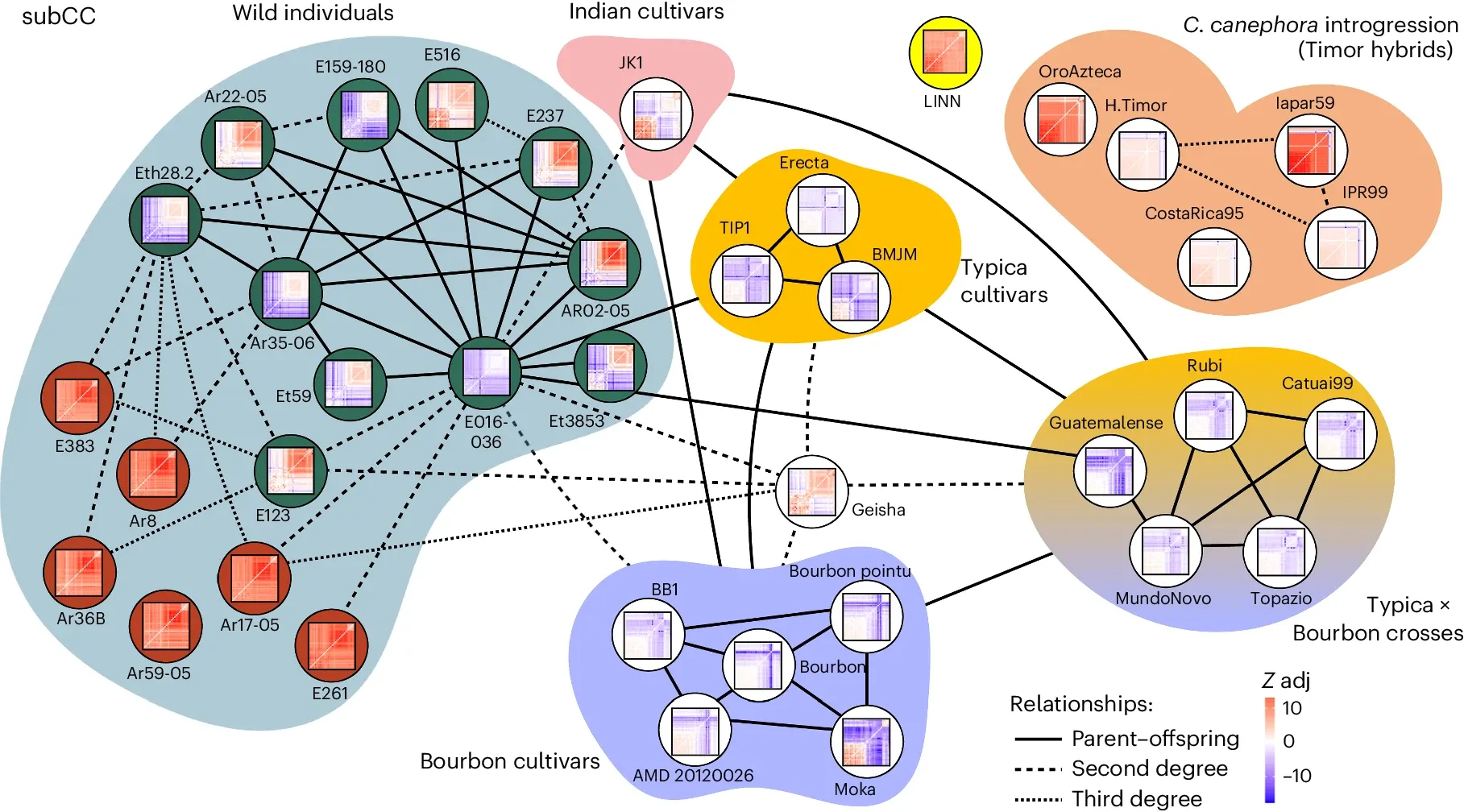
While Arabica is a natural hybrid, it has also been subjected to human selection pressures with ‘cultivars’ or varieties of Arabica being established throughout the tropics as a result of smuggling plants to places like India, Reunion Island, Timor, Asia, and the Caribbean.
We have a decent idea of how these plants made it to all of these places, but until the invention of advanced genome sequencing technologies we had little understanding of how each of these were related or how their genetics accounted for their differing traits.
Unfortunately, modern Arabica varieties exhibit low genetic diversity and are vulnerable to pests and diseases such as coffee leaf rust.
To clear up some of these questions and aid in future molecular breeding efforts to beef up this incredibly important crop, the author’s of today’s paper generated high quality reference genomes for C. arabica, C. canephora, and C. eugenioides.
They then sequenced an additional 46 varieties including 3 Canephora, 2 Eugenioides and 41 Arabica which were used to generate the relatedness map seen above.
Interestingly, this analysis identified candidate genes that may be responsible for the coffee rust resistance of the Timor variety, providing an opportunity to accelerate the development of disease resistant versions of the other varieties.
This work also adds important color to our understanding of Arabica’s evolutionary history and provides a genomic basis to help us to develop resilient coffee varieties that are adapted to our changing climate.
###
Salojärvi J, et al. 2024. The genome and population genomics of allopolyploid Coffea arabica… Nature. DOI: 10.1038/s41588-024-01695-w
What's the clinical usefulness of a microbiome? We’re still trying to figure that part out!

The microbiome is composed of all of the fungi, viruses and bacteria that live on or in an organism inclusive of their omic interactions with one another.
Most discussions around the microbiome are related to health, wellness and disease, but to actually know anything about the clinical impact of the microbiome on those things, we need to do a lot of research!
There is broad agreement that the microbiome is important, but we currently have very little knowledge about ‘cause and effect’ or how broadly we can impact the microbiome to actually influence health or prevent disease.
And understanding cause and effect is very important because that tells us whether what we’re looking at when we sample a microbiome is just the ‘result’ or if it’s something we can manipulate to do our bidding.
Current research suggests it’s a little of both!
Inflammation - A dysregulated microbiome has been associated with skin rashes and mild to severe digestive problems. Much of this appears to be linked to metabolite production, some of which can activate the immune system to create undesirable side effects in the host, but create a more desirable living environment for the microbes.
Host Metabolism - Much of human microbiome research is focused on the gut microbiome and how the gut microbiome affects our ability to properly metabolize the food that we eat. We rely on bacteria to provide certain digestive enzymes and to help produce essential things like amino acids and vitamins that are not found abundantly in our diets.
Oncology - More recently, the microbiome has been studied in the context of oncology and how the tumor microenvironment might be influenced by bacteria - both in tumorigenesis and proliferation. But there is also ongoing research in how the microbiome might influence the effectiveness of cancer treatments because of how certain microbes metabolize chemotherapy drugs or modulate the immune system to reduce the effectiveness of therapies.
Pathogenic Infections - This is probably the space where we have the best clinical evidence. We know that pathogenic bacteria can cause major problems both on our skin (think MRSA) and in our digestive tract (think H. pylori or pathogenic E. coli). When these bacteria get a foothold, they take over but there is evidence that a healthy microbiome (or fecal transfer of one for gut infections) can resist these ne'er do wells.
While the sample sizes in most of these studies is very small, we’re just at the beginning of attempting to fully understand the role of the microbiome in health and disease.
Good data and properly designed studies are the first step in allowing us to make the important clinical associations that help us identify and/or engineer bacteria to fight disease or improve our health.
Mendel first described his laws of genetic inheritance in 1865. They were promptly ignored for 35 years.
Because, let's be honest, who gives a husk about peas?!?!
It probably also didn't help that his paper was titled, 'Versuche über Pflanzenhybriden.'
Which in English translates to, 'Experiments in plant hybridization.'
While this sounds exhilarating, it belies the foundational concepts described in its pages and Mendel's work breeding peas wasn't revisited until 1900 when his results were independently rediscovered and confirmed by E. Tschermak (peas), W. Spillman (wheat) and C. Correns (peas).
H. de Vries (flowers) also independently characterized plant genetics in 1900 but had to be told that Mendel scooped him 35 years earlier.
Oof.
So, what was it that Mendel discovered while studying peas?
He observed the physical traits of peas and how these traits were passed to their offspring after breeding.
Mendel established 3 genetic principles from these observations:
Segregation - Traits come in two forms but only one from each parent is passed to offspring.
Independent Assortment - The segregation of the forms of each trait occurs independently of any other trait.
Dominance - Dominant forms of each trait mask the recessive forms and they occur in a 3:1 ratio.
Mendel initially described these as laws, but we know now that there are numerous exceptions, so in genetics we often refer to Mendelian and non-Mendelian inheritance.
But in the early 1900's, the race was on to see if Mendel's phenomenon wasn't just some weird plant thing.
One of the biggest proponents of Mendel at the time was William Bateson.
Bateson is a forgotten figure nowadays, but he popularized the works of Mendel and also did some of the earliest work on genetic linkage.
He also became fast friends with a clinician scientist, Archibald Garrod.
Garrod was most interested in the biology of the diseases in his patients and had a particular fondness for chemistry.
This could be why he was so enamored with the color of his patients’ urine.
This curiosity paid off in 1899 when he first noticed a chemical aberration in the urine of patients with Alkaptonuria.
This is an ancient disease with symptoms being described as far back as 1500 BC, but one of its tell-tale signs is darkly pigmented pee.
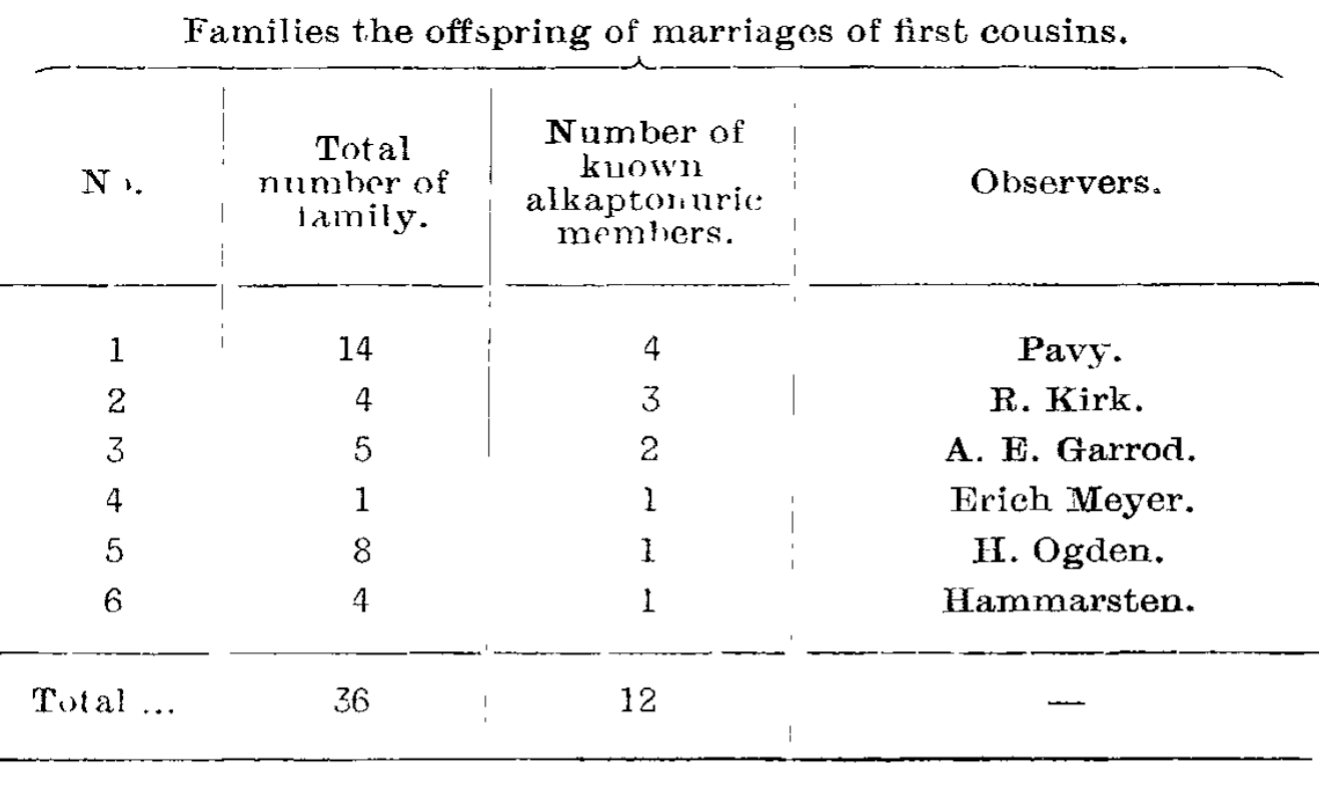
Garrod noted to his friend Bateson that this disease was found most often as the result of marriages between first cousins, and at Bateson’s urging, Garrod documented the incidence of Alkaptonuria in these families.
The table above displays the first evidence of a recessive Mendelian disease in Humans.
In it you can see that in these families the dominant form of the trait is observed in 36 offspring, and the Alkaptonuria form in 12.
This perfectly aligns with the 3:1 ratio Mendel first observed in his peas!
While this was the first description of a human Mendelian disease, we have now identified over 7,000 such diseases.
###
Garrod AE. 1902. The incidence of alkaptonuria: a study in chemical individuality. The Lancet. DOI: 10.1016/S0140-6736(01)41972-6
Were you forwarded this newsletter?
LOVE IT.
If you liked what you read, consider signing up for your own subscription here:
