Omic.ly Weekly 21
April 21, 2024
Hey There!
Thanks for spending part of your Sunday with Omic.ly!
This week's headlines include:
1) The tumor microbiome: friend or foe? Wait, there's a tumor microbiome??
2) “The microbiome” isn't just one thing. It's lots of things.
3) The structure of the DNA double-helix was published in 1953, but it took another 13 years to actually crack the genetic code.
What you missed in this week's Premium Edition:
HOT TAKE: PacBio, we love you dearly, but it’s time to step away from the short-reads
Solid tumors account for the majority of cancers that occur in adults. You might be surprised to know that they also have their own microbiome!
It’s been known for ~100 years that tumors have bacteria associated with them, but studying those bacteria has always been a bit of a challenge.
First of all, the number of bacteria associated with tumors is small and they’re mostly found inside of the tumor cells or the immune cells that infiltrate those tumors.
Secondly, tumor preservation methods make the tumor cells and anything in them non-viable.
So, capturing the bacteria and culturing them to get enough to figure out what was actually in there was basically impossible.
Fortunately, we now can just sequence these samples!
And recent studies have shown that solid tumors can contain highly complex microbial communities.
This is in addition to all of the other things we had to worry about in the tumor microenvironment!
But until now, few studies have looked comprehensively at how those microbiomes change after a primary tumor metastasizes.
It stands to reason that changing the location of a tumor could also change what microbes are associated with it!
This is important because metastases are associated with ~70% of solid tumor cancer deaths.
So, maybe there’s something we can learn about the microbiomes of these killer tumors that we can use against them as a cancer therapy?
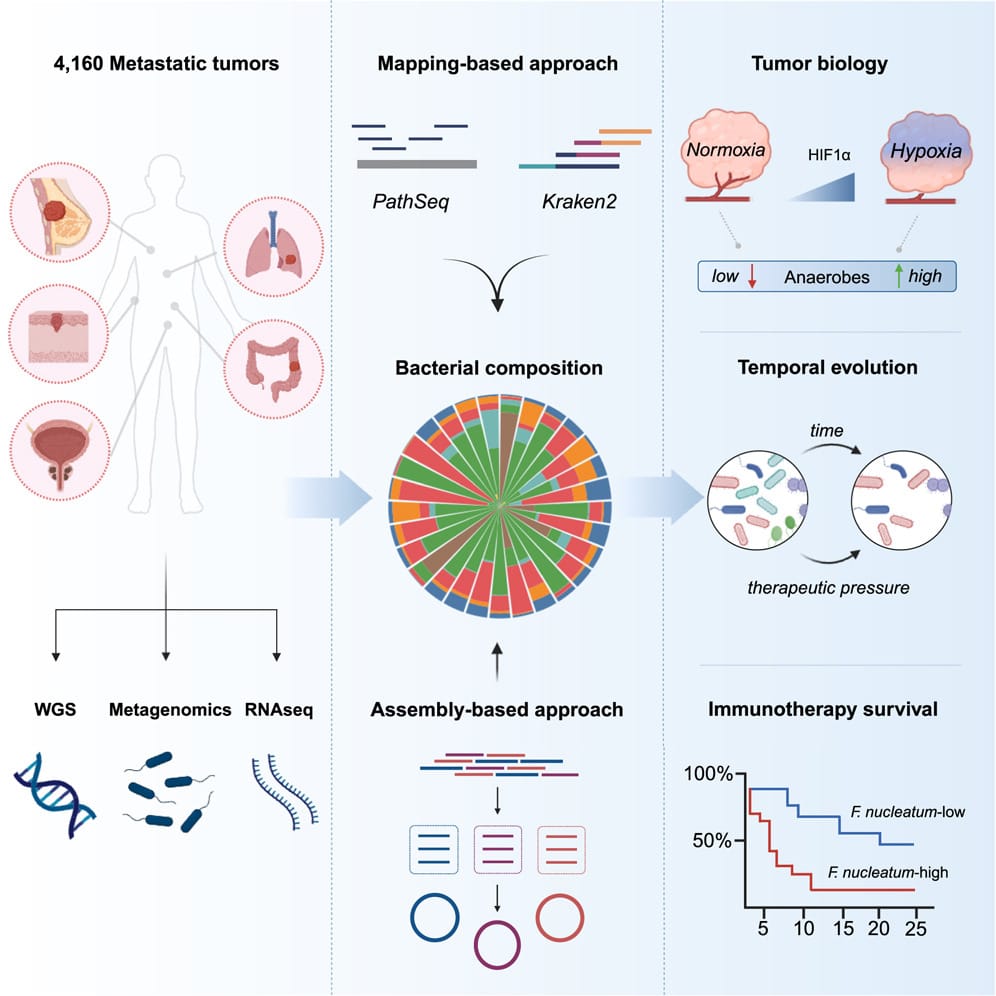
At least that’s the hope, and that’s why the authors of today’s paper chose to look at 4,160 metastatic tumor biopsies from 26 different cancers.
The findings of this study reveal that the tumor microenvironment is extraordinarily complex and that interactions between tumor cells, the immune system and the microbiome can have paradoxical roles in both promoting tumor progression and sensitizing them to therapy.
They used whole genome sequencing and whole transcriptome sequencing to perform a comprehensive metagenomic analysis to reconstruct the microbiomes of each tumor.
They showed that community composition was dependent on metastases location and that these tumors were primarily composed of anaerobes (don't need oxygen).
By comparing tumor microbiomes over time they found that these microbiomes can influence the composition of the tumor microenvironment and the microbiomes themselves can evolve after exposure to therapy.
They also showed that tumors with diverse microbial communities were more responsive to treatment and that high Fusobacterium containing metastases were more resistant to immune checkpoint blockade.
Overall, this study provides insight into the complex role of the metastatic tumor microbiome in cancer biology and therapy response.
The data are also a resource for future research on the potential therapeutic targeting of the metastatic cancer microbiome.
###
Battaglia T, et al. 2024. A pan-cancer analysis of the microbiome in metastatic cancer. Cell. DOI: 10.1016/j․cell.2024.03.021
“The microbiome” isn't just one thing. It's lots of things and each habitat has its own.

To say the human microbiome is complicated is an understatement.
It's generally agreed that a microbiome is the community of organisms (bacteria, fungi, viruses, archaea) that live and interact in a particular habitat inclusive of the environmental, physical and chemical properties of that habitat.
This means that we're surrounded by microbiomes and if we're talking about them from the human perspective, there are at least 6 that we need to discuss:
Respiratory tract - The upper airway, your nose and the cavity behind it are colonized by specific microbes that thrive in the mucosa that helps to warm, humidify and filter the air that we breathe. Similarly, the lower respiratory tract and lungs host their own crew of microbes. The microbiomes in these regions change significantly during respiratory infections or after the development of respiratory diseases like asthma.
Eye - Anyone who has ever suffered from conjunctivitis (infection of the eye) understands the importance of having a healthy eye microbiome. But the eye has three distinct habitats which include the outer eye, the conjunctiva (the membrane that covers the eye), and the meibum (the fatty, slippery, liquid that lubricates our eyes!). Each of these are inhabited by specific microbes that thrive in these ocular environments!
Oral - Our mouth is one of the microbiomes we’re most conscious of. Dental hygiene is extremely important for human health, and a dysfunctional oral microbiome is typified by unpleasant odors as a result of infections of our gums and teeth.
Skin - This is one of the more complicated microbiomes to classify since “skin” is present on every outer surface of our body. The skin microbiome can differ significantly depending on whether we’re talking about our armpit, scalp, or face! But be aware, this surface that you scrub clean every day is host to trillions of microbes!
Urogenital - This primarily refers to the vaginal microbiome which changes throughout a woman’s cycle and during pregnancy. The microbial community here is sensitive to changes in pH (and can cause changes in pH!). Dysfunction can lead to infertility or even miscarriages.
Digestive - The “gut” microbiome is probably the one that we’ve heard the most about. But the digestive tract is made up of the stomach, small intestine, large intestine, and colon. Each of these has a distinct physical and chemical environment, and you guessed it, each supports its own unique microbial community!
While we contain multitudes of microbes that form communities on and in various parts of our body, we’re still at the earliest stages of trying to tease out the cause and effect relationships here that impact human health.
While it’s pretty easy to define an unhealthy microbiome when we have an infection, identifying the factors that create and maintain a healthy one is still a huge work in progress!
The structure of the DNA double-helix was published in 1953, but it took another 13 years to actually crack the genetic code.
The two key questions after the structure of DNA was figured out were:
1) How is it copied?
2) How does it code for proteins?
The first question was answered by Meselson and Stahl in 1958. DNA is copied ‘semi-conservatively’ - the old strands serve as a template for creating the new complementary strands.
The second question wasn’t fully resolved until 1966.
There, of course, were theories about how this occurred and Francis Crick gave a seminal lecture in 1957 (published as a paper in 1958), ‘On Protein Synthesis,’ which laid out the basic formula for how he believed this all worked.
He got it mostly right.
He proposed that there must be an 'adaptor' which carries the amino acids (we now call this transfer RNA, tRNA) to the template RNA (this became messenger RNA, mRNA) to synthesize the protein chain.
But Crick also hypothesized in this lecture that the genetic code is read 3 nucleotides at a time.
It was known that there were 20 amino acids and so Crick reasoned that since there are only 4 nucleotides in DNA and RNA, the ‘code’ for each amino acid couldn’t be 2 nucleotides (4x4) because that’s only 16 combinations, so optimally, it was a 3 nucleotide code (4x4x4) because that combination gave 64 possibilities - ~3 combinations for each of the 20 amino acids.
The first indication that Crick’s hypothesis was true came in 1961 when Nirenberg and Matthaei showed that an RNA sequence containing only U’s coded for poly-phenylalanine.
Inspired by this result, Crick showed that insertion or deletion of 1 or 2 bases causes proteins to become non-functional, but 3 base mutants still produce functional protein.
The code must be a triplet!
Nirenberg and Leder followed up in 1964 with triplet codes for a handful of amino acids but they frustratingly couldn't determine all of them.
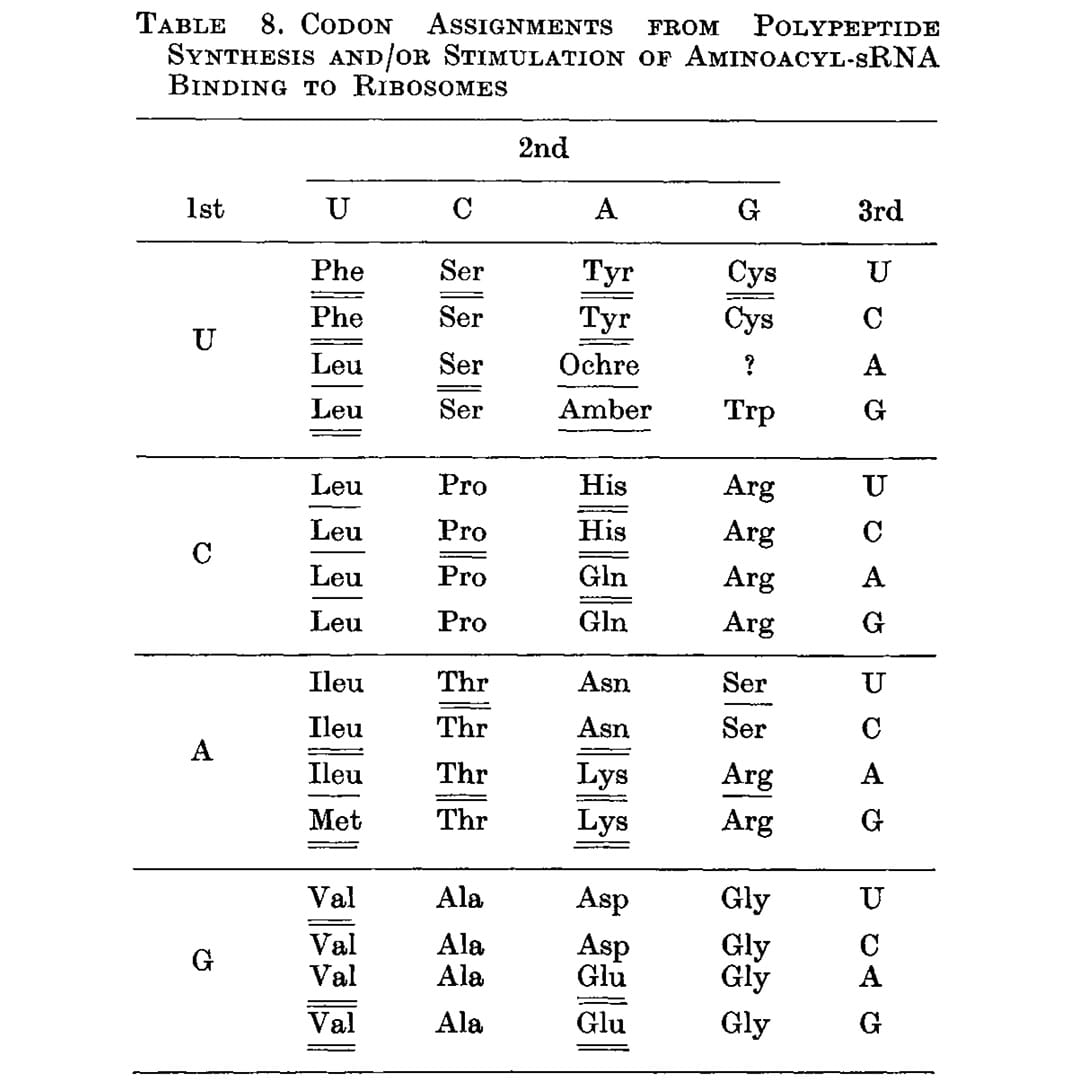
That problem was solved by Har Gobind Khorana, an Indian American scientist, whose work is the subject of the figure above.
While Nirenberg and team were using binding assays to try to capture the RNA-amino acid associations, Khorana approached it differently by synthesizing repeating dinucleotide and trinucleotide RNA polymers and seeing which amino acids ended up linked together in the resulting protein chain.
The outcome of this work can be seen below (underlines), and this is the first presentation of a nearly complete codon table. The only missing assignment is UGA, the third and final stop codon.
Unsurprisingly, Khorana, Nirenberg and Holley (he determined the structure of tRNA) shared the Nobel Prize in 1968 for deciphering how nucleic acids code for proteins.
###
Khorana HG, et al. 1966. Polynucleotide Synthesis and the Genetic Code. CSHL Symposia on Quantitative Biology. DOI:10.1101/sqb.1966․031.01․010
Were you forwarded this newsletter?
LOVE IT.
If you liked what you read, consider signing up for your own subscription here:
