Omic.ly Weekly 20
April 14, 2024
Hey There!
Welcome to the 20th issue of Omic.ly!
If you've been around since the beginning, thanks so much for your support. This newsletter wouldn't happen without you!
But since 20 is kind of a big number and we just hit 600 subscribers, I thought it would be fun to make a few changes to the website.
I've been working for the past 2 months with Arikia Millikan and her team at CTRL-X to revamp and rebrand the website using the subscription fees from paid members.
You'll still be getting the same newsletter every Sunday, but Arikia and team have been splitting up each item into their own little digestible article.
If you've found getting swamped by ~2,000 words of science every Sunday intimidating, you can now drop by the site to digest the content in a more news-y format!
So if you get some time, please stop by and check out the new page!
(We're still working out a few design bugs though!)
As always, thanks for spending part of your Sunday with Omic.ly!
This week's headlines include:
1) Colorectal cancer has found a new best friend. Spoiler: It’s a bacteria from your mouth!
2) Studying the microbiome is really just a lesson in ecology!
3) Chargaff’s Rule is the most basic rule of DNA.
What you missed in this week's Premium Edition:
HOT TAKE: Quest Diagnostics has been on a Quest to become relevant in genomics. 2024 might be the year they finally succeed.
Colorectal cancer has found a new best friend. Spoiler: It’s a bacteria from your mouth!
Our bodies host trillions of bacteria, fungi, and viruses.
A trillion is a hard number to wrap your head around but it gets even more mind blowing because we don’t have just one microbiome.
Every surface, crack and crevice creates a distinct ‘niche’ that is colonized by different microbes.
And the bugs found in your mouth are very different from the ones that hang out in your arm pits or gut!
Our microbiomes are important because, in most cases, we maintain a mutualistic relationship with these tiny invaders.
We give them food and shelter, they give us metabolites and perform chemistry that the cells in our body can’t.
But sometimes these relationships go awry and microbes that are helpful friends in one place end up being notorious enemies in another.
We’ve known for a while that the bacteria Fusobacterium nucleatum (Fn) is found predominantly in the oral cavity and is usually absent from the gut of healthy people.
However, in 2012, Fn was found co-habitating within colorectal cancer tumors.
Tumors, unsurprisingly, can also create niches, so colonization of tumors in the colon with bacteria isn’t that surprising.
But colonization with something like Fn IS because that bacteria isn’t usually found in the colon!
Why Fn bacteria is found in CRC tumors and how it gets there has been something of a mystery.
It was hypothesized that they could perform useful functions for tumors, promoting their growth, giving them the building blocks they need to be all cancer-y, and in some cases, soaking up cancer drugs and preventing them from getting to the tumor to kill it!
This mystery persisted because the sequencing tools we’ve had at our disposal haven’t been good enough to fully genetically characterize these bacteria.
Thankfully, long-read sequencing exists now and we can easily create whole genome sequences to figure out what makes them tick.
And that’s exactly what the authors of today’s paper did!
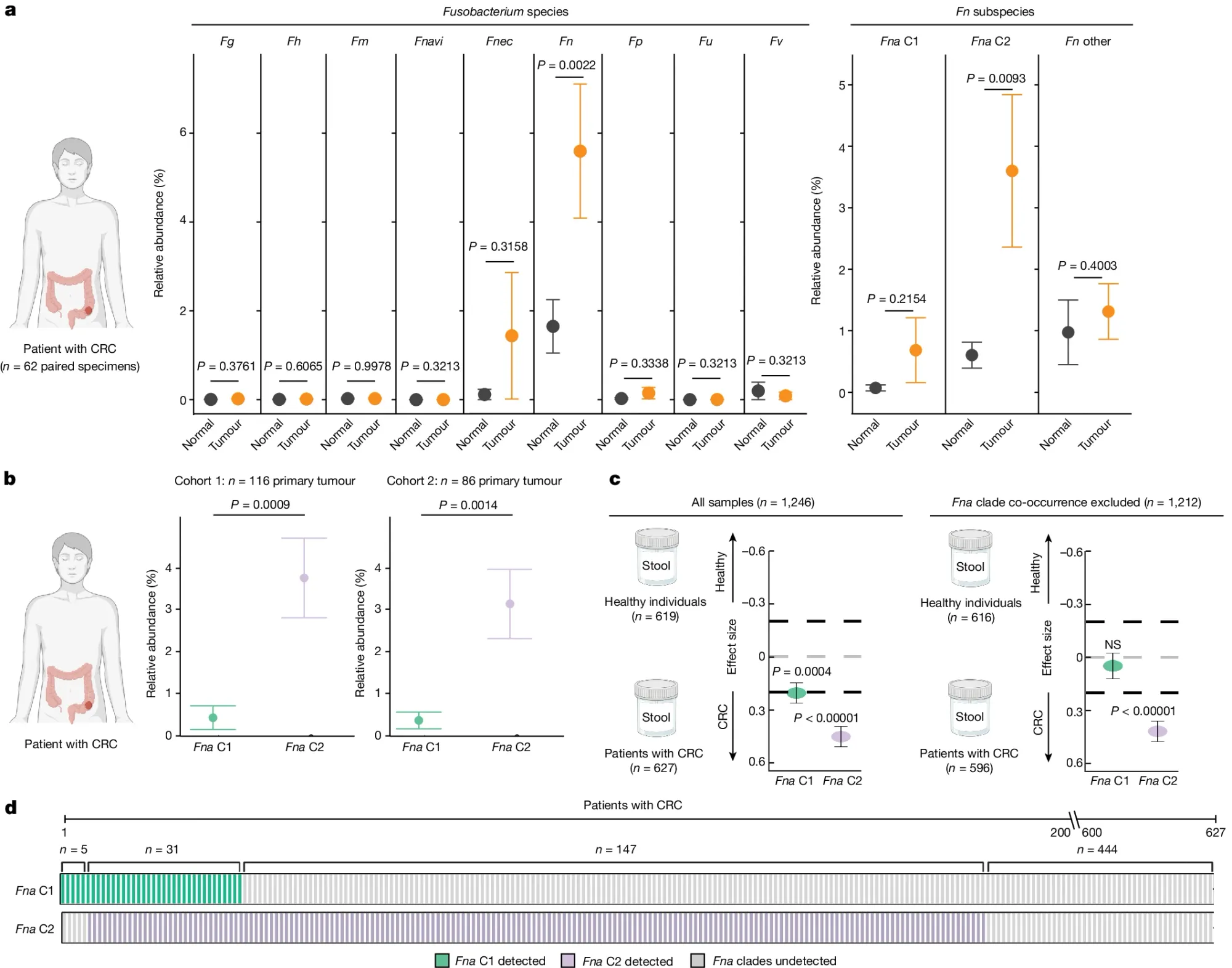
In the figure above they a) sequenced normal and CRC tissue to see which Fn subspecies were present in the gut and identified a specific sub-species of Fna, C2 (Clade 2), that was associated with CRC tumors, b) they expanded the analysis to two additional CRC cohorts showing enrichment of Fna C2 (purple) in CRC but not Fna C1 (green), c) Fna C2 is more closely associated with CRC and d) Fna C2 is present in the metagenomes of most CRC tumors.
The researchers also show that addition of Fna C2 to a mouse model of CRC promoted tumorigenesis pointing to a role for Fna C2 in increasing cancer risk and identifying it as a potential target for therapeutic and diagnostic development.
###
Zepeda-Rivera M, et al. 2024. A distinct Fusobacterium nucleatum clade dominates the colorectal cancer niche. Nature. DOI:10.1038/s41586-024-07182-w
Studying the microbiome is really just a lesson in ecology!
The word microbiome was first used to describe all of the microbes that inhabited a specific environment.
Many of the words we use when talking about the microbiome come from ecology which is the study of how organisms (usually plants and animals) interact with each other and their environments.
So, the microbiome is actually something that’s studied as a part of the field of microbial ecology which covers basically all of the different microbiomes.
We might like to think that “the microbiome” is a human centric thing, but that’s a surefire way to make a microbial ecologist angry!
Because plants, animals, the soil, and even your toilet seat have a microbiome!
In traditional ecology, a biome is a specific geographic area that is defined by the climate and the plants and animals that inhabit it.
But instead of talking about polar bears and walruses living in the arctic tundra, for the gut microbiome, we talk about the Prevotella, Bacteroides, and Clostridium that colonize your colon!
Today the microbiome is defined as the community of microorganisms that live on or in a specific environment and how they interact with one another.
These include bacteria, fungi and viruses but also the activities and interactions of their epigenomes, genomes, transcriptomes, proteomes and metabolomes.
Much like any other complex biological system, the only way to truly understand the microbiome is by looking at all of the major -omes!
This also helps us to understand what roles all of these organisms play within their community and how they survive in their niche.
Because just like in ecology, relationships exist between organisms and these can include:
Predation - Pretty straight forward, a predator eats prey.
Competition - A battle between two or more organisms over limited resources.
Commensalism - One organism benefits from a relationship and the other is unchanged.
Mutualism - Both organisms benefit from a relationship.
Parasitism - One organism benefits while the other is harmed.
So, why am I going into excruciating detail about ecology as it relates to the microbiome?
Because it's important to understand that if we want to learn anything about how we can alter the microbiome to improve human health we also need to realize that it's an incredibly complex biological system all in itself.
The microbiome isn't just about identifying the organisms in a community and saying “these are healthy and these other ones aren't.”
That's because the role of an organism changes in complex ways based on what other organisms are present and the environment in which it resides.
So, simple approaches and explanations of the microbiome won't do and there's still a lot of science that needs to be done before we can start changing the human microbiome to our benefit.
A and T, and G and C are present in the same amounts in DNA. It’s the most basic rule of DNA. It’s also called Chargaff’s Rule.
Well before the structure of the DNA double helix was revealed by Watson and Crick (with the help of Franklin and Gosling!), biochemists first had to figure out the chemical make-up of deoxyribonucleic acid.
Identifying the bases and how they were linked together with a phosphate backbone was the work of Phoebus Levene.
He was the first to tease out the chemical makeup of ‘desoxypentose nucleic acid,’ which we lovingly refer to today as DNA!
However, because only 4 bases existed, Levene triumphantly declared that DNA was far too simple to be considered the chemical that stored the genetic information of organisms.
He fell into the same trap as most everyone else at the time, and believed that the genetic material was protein.
Levene proposed the ‘tetranucleotide hypothesis’ which stated that DNA was made up of repeats of the same 4 nucleotides and these nucleotides were present in equal amounts.
We now know that this was totally wrong and part of the reason why Levene thought that nucleotides were present in equal amounts was because the analytical methods used in 1928 were not accurate enough.
It took another 22 years, and the invention of much more sensitive paper based chromatography techniques to tease out the true chemical nature of DNA.
But the first cracks in the hypothesis that ‘proteins are the genetic material’ appeared in 1944 when Avery, Macleod and McCarty showed in a series of experiments that DNA, and not protein, could ‘transform’ the bacteria pneumococcus.
While the majority of scientists at the time ignored this paper (because the results were contrary to popular belief) Erwin Chargaff of Columbia University was intrigued.
He saw Avery’s results as the first evidence that maybe the field had it all wrong about DNA, and that it wasn’t just a random jumble of nucleotides but something much more important.
So, he did what he did best, got in the lab, extracted a bunch of DNA from a bunch of different organisms and calculated in exacting detail how much of each base was present in every sample he tested.
A good number of these papers are in German, but the one that I’m pulling from today was published in Nature in 1950.
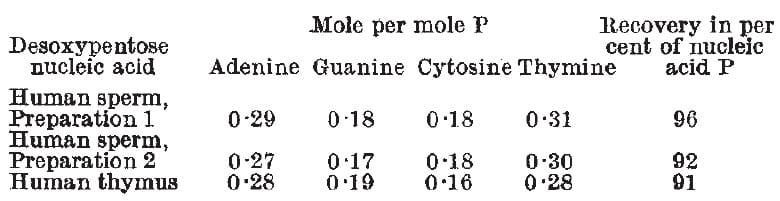
The figure above shows the nucleotide content of human sperm and human thymus.
~28% of human DNA is made up of Adenine, 18% Guanine, 18% Cytosine, and 30% Thymine.
These results show that the nucleotides aren’t all present in the same amounts as Levene had hypothesized, but that Adenine and Thymine, and Guanine and Cytosine are found in equal concentrations.
This was a key discovery on the path to showing how these seemingly complementary nucleotides bound together in the structure of DNA.
###
Chargaff E, et al. 1950. Composition of human desoxypentose nucleic acid. Nature. DOI:10.1038/165756b0
Were you forwarded this newsletter?
LOVE IT.
If you liked what you read, consider signing up for your own subscription here:
