Omic.ly Weekly 19
April 7, 2024
Hey There!
Thanks for spending part of your Sunday with Omic.ly!
This week's headlines include:
1) Sometimes you gotta break a couple DNA strands to make a memory.
2) So, you want to learn more about your microbiome?
3) Deoxyribonucleic acid (DNA) is the genetic material, but early on, most of science thought the genetic material was protein. That changed in 1952.
What you missed in this week's Premium Edition:
HOT-TAKE: Illumina sheds a board Icahn and, hopefully, most of the drama that came with it.
Sometimes you gotta break a couple DNA strands to make a memory.
Or at least that's what recent research into memory formation in mice suggests!
The brain is the most complex organ in the body and so it shouldn't come as a surprise that memory formation is also extremely complicated!
There are three major types of memory:
Sensory - Memory perceived after viewing (Iconic), touching (haptic) or hearing (ehoic) something. This memory fades after a few hundred milliseconds.
Short-Term - Memory that lasts a few seconds to minutes, but has a limited capacity, and most people can only remember 4-5 items stored in short-term memory.
Long-Term - Memory that can store large amounts of information for long periods of time.
How these different types of memory work has been a hot topic of debate since the early 1900’s.
But sensory and short-term memory are believed to be the result of short lived connections between neurons in the frontal lobe.
Conversely, long-term memory is formed from permanent connections between neurons throughout the brain and, just in the last decade, we have begun to understand how this happens on the molecular level.
It's been linked to changes in DNA methylation and gene expression and this link was discovered by comparing the brains of rodents before and after the formation of a long-term memory.
But, how does one go about creating long-term memories in rodents?
By changing their environment and then shocking them.
This is called ‘contextual fear conditioning’ (CFC) and the hallmark of CFC is the freeze response.
Not surprisingly, after a traumatic experience, rodents store this information in their long-term memory and freeze when placed in that situation again.
The authors of today's paper used CFC to study the molecular changes that happen in neurons before, immediately after and a couple months after conditioning.
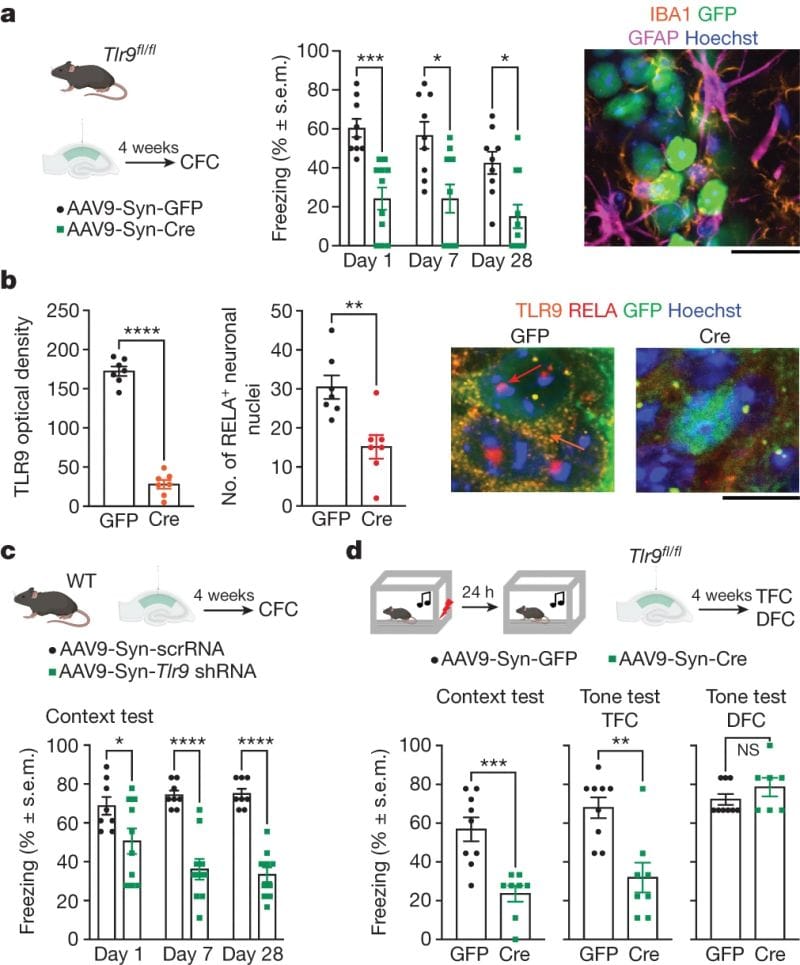
What they saw was that memory forming CA1 neurons ramped up expression of immune response genes but most notably TLR9 and the NF-κB signalling pathway.
TLR9 detects the presence of double stranded DNA, usually as a defense against bacterial infection, but the authors hypothesized that causing DNA breaks was important in the formation of long-term memories.
To test this they created mice where they could selectively turn off TLR9 expression.
The figure below shows the results of this experiment: a,c) treated (green), but not control (black), show a reduced freeze response. b) TLR9 and its signaling partner RELA are significantly reduced after treatment.
In another experiment, DNA breaks were also shown to increase after CFC.
These results indicate that TLR9 and DNA breaks play a role in memory formation and could be an important therapeutic target in neurodegenerative diseases.
###
Jovasevic V, et al. 2024. Formation of memory assemblies through the DNA-sensing TLR9 pathway. Nature. DOI: 10.1038/s41586-024-07220-7
So, you want to learn more about your microbiome?
Cool, here's what you need to know!
The human microbiome is the entire community of microorganisms that live on or in your body and is made up of bacteria, fungi and viruses.
Your body contains, or has on it, many different microbiomes.
This is because each crack, crevice, surface, and orifice creates a distinct environmental niche that promotes the growth of totally different microorganisms!
Meaning, your skin, your mouth, your armpits, and your gut have different microbiomes.
But it doesn't stop there, your gut microbiome itself can be segmented into different microbiomes because your upper and lower gastrointestinal tract create completely different environments.
So If you want to learn about your microbiome, you need to pick one, or choose a service that gives you multiple swabs to test them all!
Well, all except your upper GI, that one's a bit tough to do yourself, even if you have a reeeeallllly long swab.
Once you've settled on which one you want to test you need to pick a service provider.
This is a pretty crowded space but I'm going to let you in on a little secret.
Most of these tests are actually just a resold product from a handful of vendors who provide 'white label' services, so you're mostly getting the same thing from everyone.
And what you're generally getting is a read-out of your microbial community based on 16s ribosomal sequencing.
Now, only bacteria contain a 16s gene so the vast majority of services aren't really reporting out a true microbiome because they're missing all the fungi and viruses!
But to make matters worse, most of these services only sequence a small fraction of the 16s gene, the V4 hypervariable region.
You might be thinking, 'hypervariable sounds good!'
Except the 16s gene is 1600 bases long and has 9 hypervariable regions...
Sequencing just one of them is not going to give you the most accurate representation of your community.
And if you really want to do this right, you'd do 16s-ITS-23s.
Except you can't sequence that unless you use **drum roll** long-reads because it's ~4,500bp!
Microbiome sequencing is yet another example of a situation where 'friends don't let friends short-read,' especially if you actually care about identifying species or subspecies level community composition.
There are products that claim they can get species level descrimination from just V4.
But don't be fooled!
While they can detect about 50% of species with just V4, you really need all 9 hypervariable regions to get a species level identification for all of the taxa in a sample.
Otherwise, the best you can do is a genus level identification.
So, what do you actually get out of doing a direct to consumer microbiome analysis?
That's a good question.
Because there's general agreement that the microbiome is important, but what we can actually learn by sequencing it, other than saying there's some bacteria there, is still a work in progress.
Deoxyribonucleic acid (DNA) is the genetic material, but early on, most of science thought the genetic material was protein. That changed in 1952.
Ok, it didn’t change overnight, because scientists are pretty stubborn.
It actually wasn’t accepted until the 60’s, but, in 1951, Alfred Hershey and Martha Chase began a series of experiments that conclusively showed that DNA was the genetic material.
There was already good evidence that the genetic material wasn’t protein, but early molecular biologists couldn’t wrap their heads around how 4 bases of DNA could code for all of the diversity we see, especially when proteins are made up of long chains of 20 amino acids.
Surely, since 20 is greater than 4, you get more diversity with 20!
So the hypothesis was that DNA must be the scaffold that holds everything together.
Of course, we know now that’s absolutely not true, and it’s basically the opposite - DNA is the genetic material and protein serves a support function.
The work that synthesized all of these ideas together was completed by Hershey and Chase in what are now referred to as the Hershey-Chase Experiments.
They worked with bacterial viruses called bacteriophages.
These are very simple viruses that consist of a protein shell surrounding the DNA of the virus.
But before the 1950’s, no one knew what phages looked like or how they infected bacteria.
In 1951, Thomas F. Anderson, a pioneer of electron microscopy, showed that phages are like little moon landers and attach themselves to the outside of the bacteria but do not enter them.
This created a bit of a conundrum, how can protein be the genetic material if none of the phage protein gets inside?
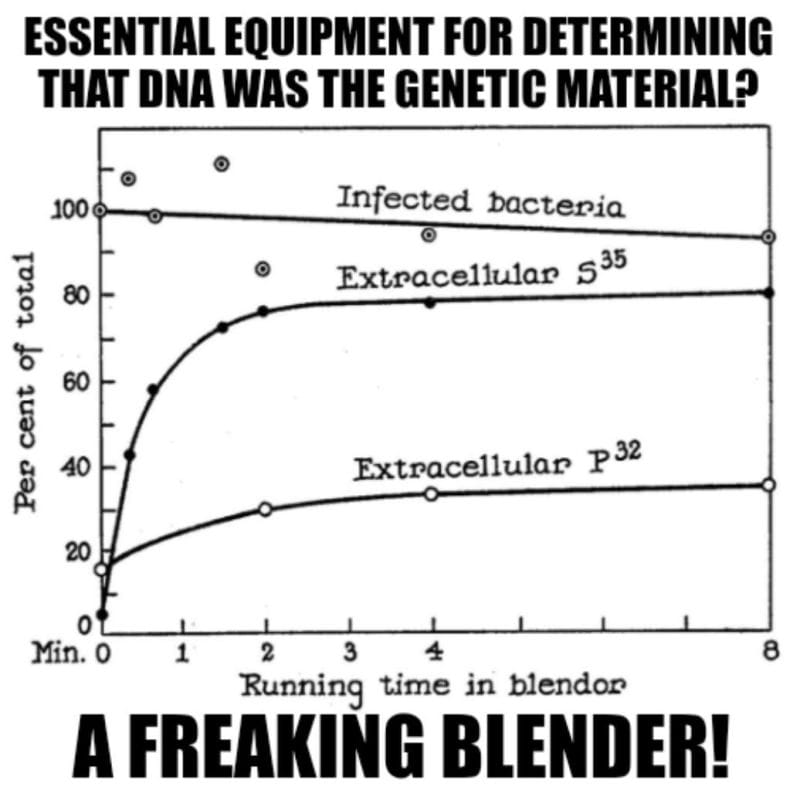
The answer to this question involved Hershey-Chase, a Waring blender and phages grown in the presence of radioactive food items.
Luckily, proteins do not contain Phosphorus (P) and DNA does not contain Sulfur (S), so radioactive P-32 and S-35 could be used to label each component of the bacteriophage individually.
Radioactive phages were then combined with bacteria and stuck in a blender.
Hershey and Chase discovered that this technique didn’t kill the bacteria, but it was perfect for shaking the empty phage husks from the bacteria.
This is documented in the figure below which shows 90% viability of the infected bacteria but that 80% of the protein (S35) remained external while only 30% of the DNA (P32) does after blending.
This result supported the electron microscopy observation that the phages are basically just little protein covered DNA injectors!
Hershey and Chase followed up by showing that 30% of the DNA that makes it into baby phages after the blender experiment was radiolabeled while <1% of the protein was, essentially proving that DNA, and not protein, is the heritable genetic material.
###
Hershey, AD and Chase, M. 1952. Independent Functions of Viral Protein and Nucleic Acid in Growth of Bacteriophage. J Gen Physiol. DOI: 10.1085/jgp.36.1.39
