Omic.ly Weekly 18
March 31st, 2024
Hey There!
Thanks for spending part of your Sunday with Omic.ly!
This week's headlines include:
1) Human fetal brain organoids: a controversial new platform.
2) Genetic code expansion and how we get to designer proteins.
3) Commercial industrial engineering is the savior of high-throughput genomics.
What you missed in this week's Premium Edition:
HOT-TAKE: The holy grail of blood based cancer early detection still can't beat feces when it comes to colorectal cancer screening.
Human fetal brain organoids: a glimpse into the developing brain and a platform for future discovery
The brain is the most complicated organ in the human body.
And the human brain is probably the most complicated organ of any species on earth.
It has a highly folded and layered structure with much greater cellular diversity than the brains of other mammals.
It's also the organ that gives us our intelligence and basically defines us as humans.
We've learned a lot about the brain by studying it extensively in animal model systems.
In the last few decades we've also discovered how to study organ tissues and their development by isolating and culturing stem cells outside of the body.
And more recently we've figured out how to grow these cells into 3D structures called ‘organoids’ which can mimic the cellular behavior of organs.
For example heart organoids can beat and some brain organoids can have electrical activity that allow us to study in more detail how these organs develop and function.
But stem cells can be a pretty controversial topic because of how they are obtained.
They come in two forms, tissue stem cells (TSCs) and pluripotent stem cells (PSCs).
TSCs are cells from tissues that when grown in the right conditions maintain their cellular identity.
The easiest TSCs to grow are from tissues that have an innate capacity to regenerate like intestinal or liver cells.
PSCs are cells that can basically turn into any cell in the body. They haven't differentiated yet and under the right conditions can be coaxed to form a wide variety of different cell types.
Some TSCs can be derived from adult tissue, and adult cells can also be reprogrammed into something that looks and behaves similarly to a PSC, but the best source for each of these stem cell types is to obtain them from fetuses.
That's because fetal tissues behave very differently from adult tissues and fetal cells can be cultured and expanded for long periods of time before they become unusable.
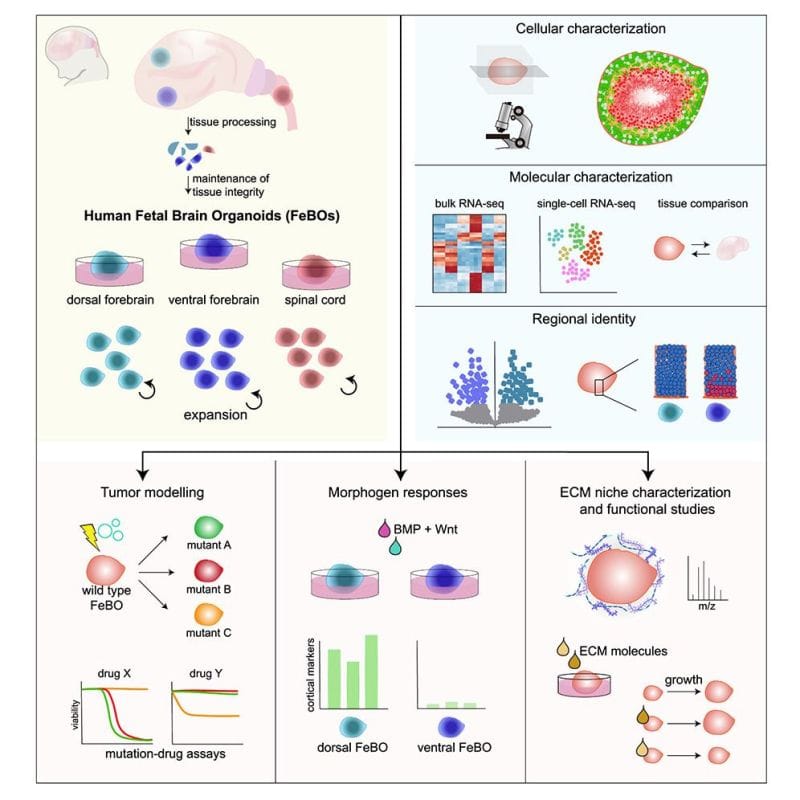
The authors of today's paper were the first to establish stable cultures of human fetal cells from 3 different regions of the central nervous system: dorsal forebrain, central forebrain and spinal cord.
They showed that these distinct cellular populations could maintain their identity indefinitely and would only differentiate into more mature compositions when switched to a maturation media.
These fetal cells also self-organized into complex organoids (FeBOs), forming more natural cellular microenvironments including the establishment of an extracellular matrix.
These are important developments because these organoids can be used for studying the brain's cellular microenvironment, brain development, or as models for brain tumors and their treatment.
###
Hendriks D et al. 2024. Human fetal brain self-organizes into long-term expanding organoids. Cell. DOI: 10.1016/j․cell․2023.12.012
Genetic code expansion might be the coolest thing in Omics that you've never heard of.

DNA codes for messenger RNA (mRNA) which is then translated into protein.
And it's proteins that perform the majority of the functions in our cells!
They can serve as structural components, as molecular motors delivering cargo to various regions of the cell, and they can function as enzymes performing chemistry that would otherwise be impossible.
Proteins are created by ribosomes which read mRNA and stitch together amino acids to form the final protein sequence.
They do this by binding to mRNA and transfer RNAs (tRNAs) that are loaded with specific amino acids.
The sequence of the amino acids in a protein is determined by the mRNA sequence (codon) binding to a complementary tRNA (anticodon).
Proteins are made using 20 common amino acids but there are over 500 of them found in nature!
Many of those extra amino acids are just modified common amino acids, but this presents an interesting challenge:
What if we could expand the number of amino acids that can be coded for by DNA??
Doing this would allow us to engineer totally new proteins!
And this is the focus of an exciting area of synthetic biology referred to as ‘genetic code expansion.’
We can get to these designer proteins through a number of different biological hacks.
One of them is to change what the natural base pairs (NBPs) code for!
There are 4 DNA bases (A, T, C, G), these are read 3 at a time to code for proteins and this ‘triplet code’ is referred to as a codon. There are 64 potential codons and 20 common amino acids. Each amino acid is coded for by approximately 3 different codons.
But, we have the tools to change that!
Stop Codons - Are triplet codes that don’t code for any amino acids but serve to tell a ribosome to stop; however, we can make tRNAs that actually bind to the stop codons and cause a ribosome to insert whatever amino acid is attached to that stop-codon-binding-tRNA!
4 Base Codons - We can create tRNAs that use 4 base codons instead of 3 base codons (but this can get tricky in a living organism!)
Codon/tRNA Reprogramming - We can engineer a cell to incorporate a different amino acid on a specific tRNA.
But, hacking what the natural base pairs code for can have unintended consequences in the proteins we need to keep cells alive!
Thankfully, synthetic biologists have also been working to add unnatural base pairs (UBPs) to these systems and have been successful in creating organisms that can use 6! base pairs (A, T, C, G, X, and Y).
This means we can create our funky new proteins without affecting how other critical proteins are made, because we can make tRNAs that complement new triplet codes and recognize our unnatural X and Y bases.
Ultimately, this allows us to create new proteins that have desirable functions.
Genetic code expansion has important applications in academic research, protein and enzyme engineering, and therapeutic development.
We ‘completed’ the human genome in 2003, thanks mostly to commercial industrial engineering.
Genome sequencing didn’t really enter the household vernacular until the announcement of the Human Genome Project in the early 90’s, and that was only possible because of major advancements in sequencing technology.
For the better part of a decade, Sanger sequencing was performed using radiolabeled nucleotides and chain terminators, one for each base, requiring a lane for each A, C, T, and G.
In 1987, George Trainor and a team at DuPont got the bright idea, literally, to use fluorescently labeled chain-terminators, enabling the multiplexing of the Sanger sequencing reaction into a single tube.
This might sound like a simple and obvious feat, but it wasn’t!
The advancement here isn’t just “add fluorescently tagged bases and BOOM! Here’s the data.”
They had to find a compatible polymerase, synthesize the dyes, tag the nucleotides, and engineer an optical system for data acquisition.
The system itself is an engineering wonder: there’s a laser, a beam expander, a suite of scanning optics, two detectors with their own filter stacks, all aligned to excite and detect the dyes in the sequencing gel bands as they pass across a laser beam.
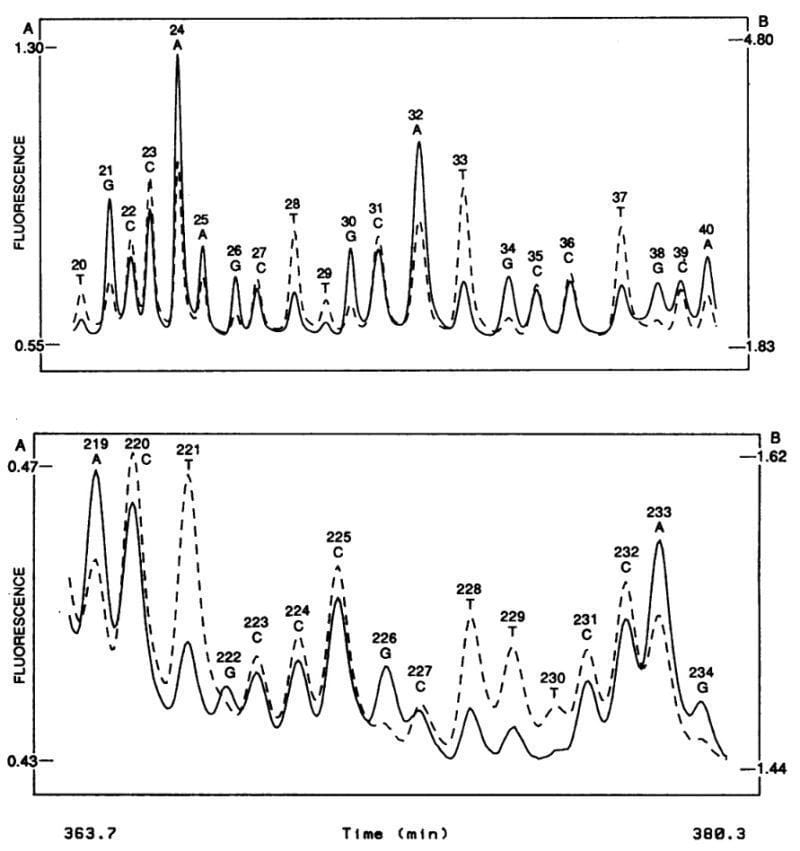
The figure above shows what this groundbreaking system was able to accomplish - the sequence of two 20 base pair stretches of M13mp18, a lac phage vector.
The solid line is the fluorescence intensity observed by the A, G detector and the dashed line the intensity observed by the C, T detector.
These plots should look suspiciously familiar to anyone that’s done fluorescence based Sanger sequencing and it probably isn’t a surprise that, through a licensing agreement, the principles of the technology in this paper made their way into the first four color commercial sequencer in 1988, the ABI 370A.
Despite this process remaining gel based: multiplexing the reaction, eliminating the radioactive nucleotides, ditching the film, and automating the acquisition of the sequencing data were all a huge triumph.
But because it still required the pouring of a gel, it gave grad students and postdocs something to complain about for at least another decade; unfortunately for them, the capillary electrophoresis based ABI Prism line wasn't introduced until 1996.
###
Prober, JM, et al. 1987. A system for rapid DNA sequencing with fluorescent chain-terminating dideoxynucleotides. Science. DOI: 10.1126/science.2443975
Were you forwarded this newsletter?
LOVE IT.
If you liked what you read, consider signing up for your own subscription here:
