Omic.ly Weekly 17
March 24th, 2024
Hey There!
Thanks for spending part of your Sunday with Omic.ly!
This week's headlines include:
1) Tumor immune microenvironments and how we can exploit them to fight cancer.
2) Proteomics: It’s all about the structures.
3) 6LL3 showed that mammalian cloning was possible in 1997.
4) Weekly Reading List
What you missed in this week's Premium Edition:
HOT-TAKE: Bionano Genomics is mapping a new course for an uncertain future
Tumors are bad. Understanding their microenvironment is key in our effort to amp up the immune system to fight them!
Cancers are a terrible scourge on humanity.
We’ve all experienced personal losses to them, but our colloquial discussions of ‘cancer’ are overly simplistic.
‘Why haven’t we cured cancer?’
Mostly because it's not one thing.
It’s thousands, maybe millions, of things.
And treatments that are effective in even the same type of cancer aren’t effective for everyone with that type of cancer!
That’s because cancers are ‘heterogeneous’ which is a big scientific word for ‘stupidly complicated.’
And cancers are some of the most stupidly complicated diseases that we have to deal with in science.
But more recently we’ve discovered how our immune system can be used to uncomplicate things.
It’s the first line of defense when cells start misbehaving, but this defense can get exhausted and even blocked by tumor cells as they evolve mechanisms to evade detection and destruction!
One important example here is the action of the proteins PD-1 and PD-L1.
PD-1 is found on the surface of immune cells and binds to a receptor, PD-L1, that sits on the outside of non-immune cells.
This tells the immune cells to not kill them.
Mostly because you don’t want the immune system going around and killing the good guys!
Eventually, tumor cells figure out this trick too and evade the immune system by producing a bunch of PD-L1.
But because we discovered this interaction, we were able to develop one of the first cancer immunotherapies, Keytruda, to take back control of the immune system and attack these resistant cancers.
If Keytruda is around, it binds to PD-1 and blocks its interaction with PD-L1 allowing immune cells to commence the killing!
This has been a revolutionary drug to combat susceptible cancers, but what if there were other ways we could get around tumor immunity to attack cancers?
That’s where today’s paper comes in!
Because you can’t destroy tumors if you don’t know their weak points.
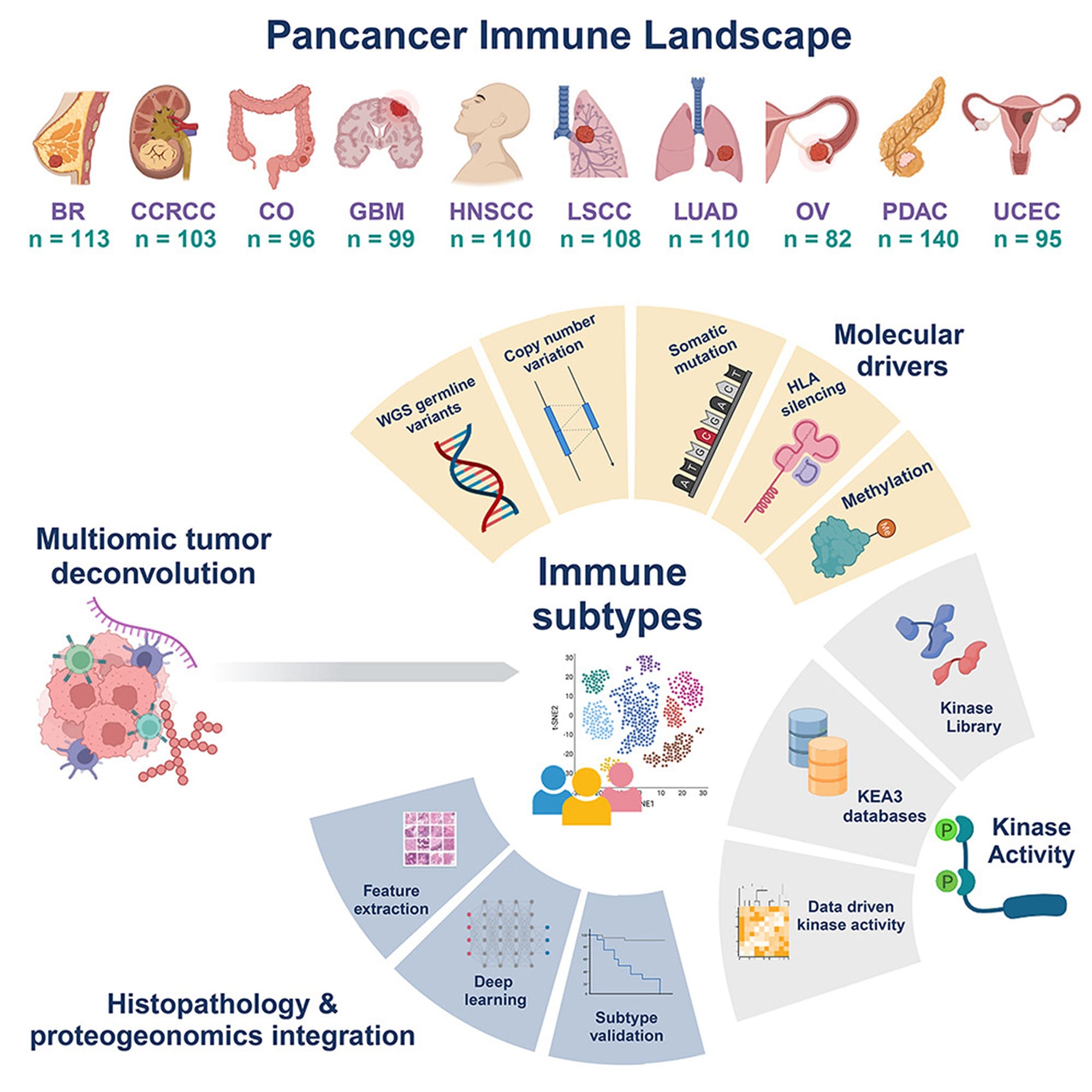
And to find those weak points, the authors cast a very wide net (see image below) looking at genetic, epigenetic, spatial transcriptomic, spatial proteomic and phosphoproteomic (kinases!) data in breast cancer (BC), renal carcinoma (CCRCC), colon cancer (CO), glioblastoma (GBM), head and neck cancer (HNSCC), two lung cancers (LSCC and LUAD), ovarian cancer (OV), pancreatic cancer (PDAC), and uterine cancer (UCEC).
There's a boat-load of information in here, and that’s not surprising since it’s a product of the Clinical Proteomic Tumor Analysis Consortium!
But what makes this dataset unique is it comprehensively profiles the tumor immune microenvironment, opening up new possibilities for targeted therapeutics based on the 'immune subtype' of each individual tumor.
###
Petralia F et al. 2024. Pan-cancer proteogenomics characterization of tumor immunity. Cell. DOI: 10.1016/j․cell.2024.01.027
Proteomics: It’s all about the structures!
‘Structure Equals Function.’
It’s basically the secret password at the molecular biology club.
And if you’re ever looking to get knowing nods from your favorite molecularly aligned friends, this phrase kills.
That’s because most everything in molecular biology is centered on how molecules interact with one another.
This includes how two strands of DNA coil into a right-handed helix, how proteins bind to one another to create functional complexes, or how enzymes grab on to their co-factors and substrates to catalyze life-sustaining chemistry.
All of these things are essentially determined by their molecular structures!
These structures are pretty easy to determine for run of the mill chemical compounds, but proteins are macromolecules.
This just means they’re composed of thousands of atoms and can have highly complex structures!
But the complexity for proteins doesn’t end there, because they can also be modified after they are made.
These ‘post-translational modifications,’ or PTMs for short, can be as simple as the addition of a phosphate to a specific part of a protein all of the way to the addition of large chains of sugar molecules.
And you guessed it, the addition of these PTMs changes the structure of the protein which in turn changes its function!
Many PTMs are critical for proteins to operate correctly and they routinely serve as ‘on’ and ‘off’ switches.
They’re also important for determining which partners a protein can interact with and when!
So, a protein’s structure is REALLY freakin’ important for understanding what it does.
'But how do we determine the structure of a protein?'
I'm glad you asked!
X-ray Crystallography - The OG of structure determination involves creating pure extracts of the protein you want to look at and finding the perfect conditions (salt concentration, humidity, drying time) to create a pristine crystal. Those crystals are then bombarded with X-rays. And the structure of the underlying protein can be inferred based on how those X-rays bounced off of it! But one major drawback of crystallography is that crystals don’t have water! So they may not necessarily represent the ACTUAL structure we’d see inside of a wet environment.
Nuclear Magnetic Resonance Spectroscopy (NMR) - Best used on small proteins and peptides. Purified protein is exposed to radio waves and its structure can be determined by looking at resonance signals and calculating which atoms within a protein are near one another.
Cryogenic Electron Microscopy (CryoEM) - Purified proteins or large protein complexes are cryogenically frozen and then imaged with an electron microscope. The images of thousands of molecules are then combined to stitch together a 3D structure of the protein!
AlphaFold - Kidding, this is a structure prediction tool. It’s important to validate what this produces with one of the above methods.
6LL3, better known as Dolly the sheep, showed that mammalian cloning was possible in 1997.
So, why was a sheep the first choice for this ground-breaking work?
Well, they’re cheap, and Ian Wilmut and his team at the Roslin Institute in Scotland were interested in creating a mammalian platform for the development of biological pharmaceuticals to treat human disease.
The first step of that pioneering vision was to show that it was possible to clone a mammal!
Dolly was created by transferring the nucleus (contains the DNA!) of a mammary epithelial cell from a 6-year old Finn Dorsett ewe into an Oocyte (fancy term for an egg) of a Scottish Black Face ewe that had been enucleated (had its nucleus removed).
The egg with the new nucleus was then implanted in a surrogate and Dolly was born!
Basically, Dolly is the 'clone' of the 6-year old Finn Dorsett sheep.
Although the authors do not use the word 'clone' anywhere at all in the original article.
But you may be wondering why this is such a big deal.
And the answer to that is because it’s really hard to clone animals!
Cells in adults have undergone differentiation – meaning, parts of their genomes have been deactivated so that the cells only produce the proteins required for their specific function within the body.
The researchers tricked these cells into removing all these deactivation marks and then inserted the 'reprogrammed' nuclei into Oocytes to see if they could create live lambs.
Technically, 8 lambs were reported in this paper, but Dolly was the only one that was derived from adult tissue.
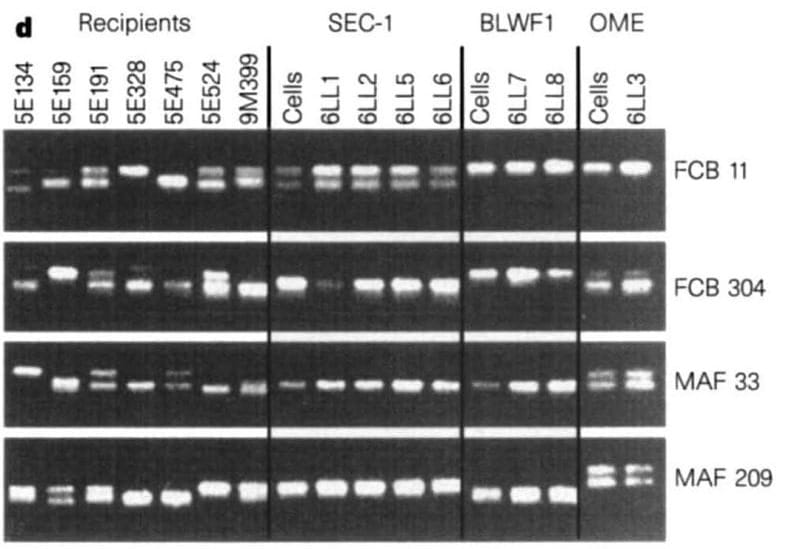
The figure above shows the microsatellite markers of the recipient ewes (the surrogates), the cells (embryo derived – SEC1, fetal-derived – BLW1, and mammary-derived – OME), and the lambs that were born.
6LL3 shares the same microsatellite markers as the mammary cells she was derived from, but not the same microsatellite markers of any of the surrogate ewes.
Dolly is a bonafide copy!
And it should come as no surprise that the popular reporting on Dolly at the time highlighted how close humanity was to creating designer babies.
Or, that scientists should stop playing god.
But this work, along with earlier work in amphibians by John Gurdon, fueled the later discovery of induced pluripotent stem cells (iPSC) by Shinya Yamanaka and led to his subsequent Nobel Prize in 2012.
The scientific value of cellular reprogramming and proof that it could be done in mammals (Dolly!) was a tremendous first step in using this and similar techniques to help us understand the complexities of our genomes, how they function, and, ultimately, the usefulness of cloning (and stem cells!) for the development of treatments for human disease.
###
Wilmut I et al. 1997. Viable offspring derived from fetal and adult mammalian cells. Nature. DOI: 10.1038/385810a0
Were you forwarded this newsletter?
LOVE IT.
If you liked what you read, consider signing up for your own subscription here:
