Omic.ly Weekly 16
March 17th, 2024
Hey There!
Thanks for spending part of your Sunday with Omic.ly!
This week's headlines include:
1) Epigenome editors are coming for cholesterol (and other diseases)!
2) PROTACs and LYTACs: How therapeutics can take advantage of proteomic garbage men.
3) One of the most important papers in the history of genetics was basically ignored when it was published in 1944.
What you missed in this week's Premium Edition:
HOT-TAKE: NanoString steps back from the ledge and looks to find new life as a private company.
Ever heard of an ‘epigenome editor?’ Well, buckle up, because they could be the source of a long-term cure for high cholesterol!
You probably know about CRISPR-Cas, zinc finger finger proteins (ZFPs), or transcription activator-like effector nucleases (TALENs) which can be engineered to bind to and directly edit DNA.
These all have been used with varying degrees of success and result in permanent changes to the DNA of the cells in which they operate.
A safer method of genome editing in many cases would be if we could flip a little switch to turn off the things that don’t work, and/or turn on the things that do!
This is where epigenome editors come in and they’re the cool new kids on the genome editing block.
They’ve been engineered using the DNA binding trick of those other editors but combined with pieces of different proteins to create protein Frankensteins that are able to turn genes on or off.
This works through epigenetic modification of DNA!
And epigenome editors are designed to attract methylases (off switch) or acetylases (on switch) to ‘edit’ how specific genes are expressed.
The nice thing about this scheme is that these changes can be reversed which could make them safer to use than their DNA slicing and dicing cousins.
As a proof-of-concept, the authors show that epigenome editors can be used to reduce cholesterol.
Their target gene of choice was PCSK9.
This is an enzyme involved in destroying the LDL receptor which pulls bad cholesterol out of the blood stream.
Getting rid of PCSK9 has been shown to be very efficient at reducing cholesterol and two extremely effective (and expensive) biologic drugs are available that do just that!
But, a long-term cure might be the preferable solution.
So, the authors engineered 3 different proteins to epigenetically modify the PCSK9 gene, two of them based on ZFPs (Engineered Transcriptional Repressors - ETR) and one based on a deactivated Cas9.
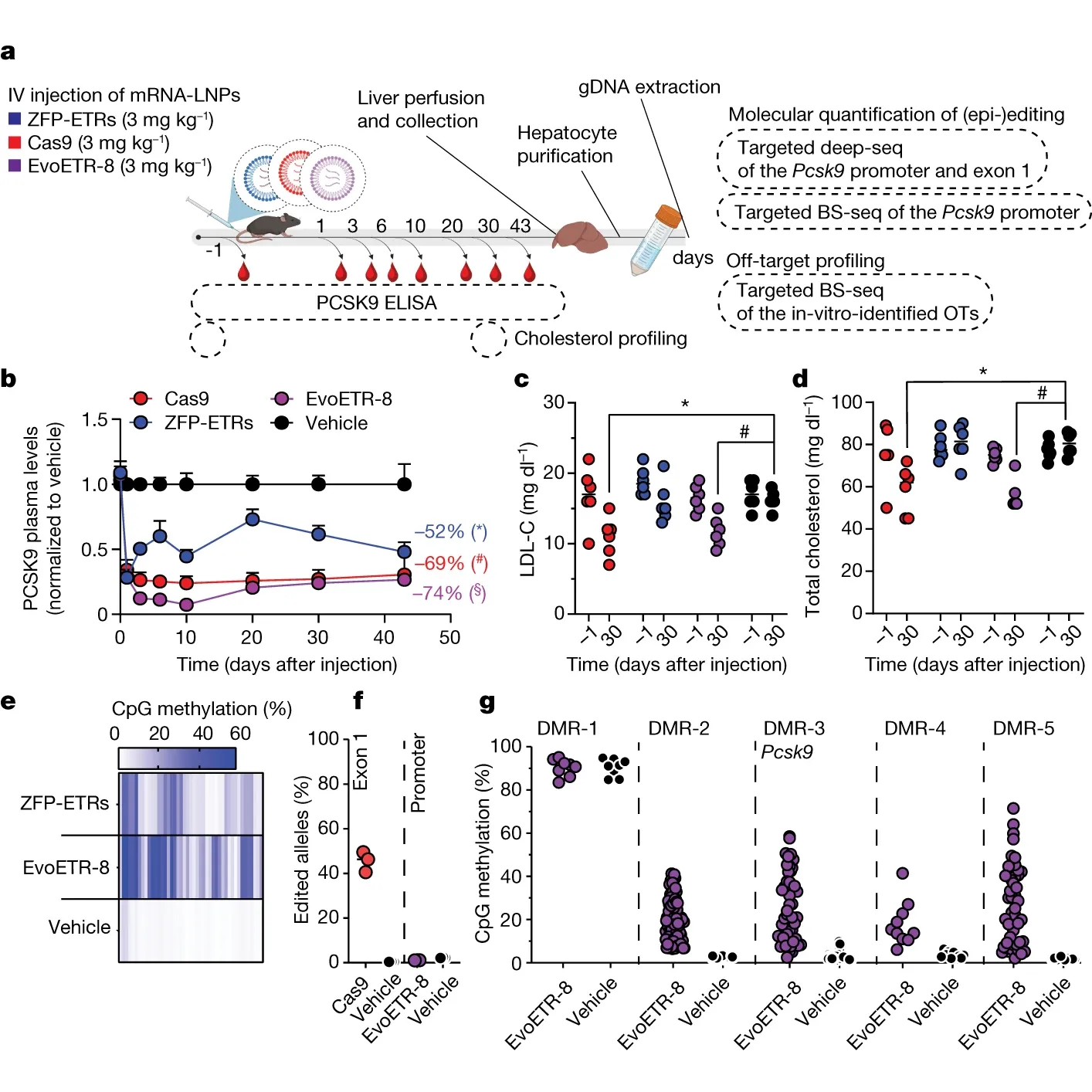
The results of this experiment can be seen in the figure above: a) shows an overview of the experimental design; b) the effect of the engineered ETRs and Cas9 on PCSK9 levels; c and d) the effect on cholesterol levels; e, f, and g) the amount of methylation in the specified regions of the genome.
While they were not able to completely eliminate the expression of PCSK9 with any of these methods, they did show that the ~70% reduction of PCSK9 and cholesterol was long lasting (100+ days), including after a partial hepatectomy, meaning the methylation signal was passed on to new cells after liver regeneration!
There’s still a bit of work left to do here before we see these solutions in the clinic, but epigenetic editors seem like they might be something to keep an eye on!
###
Cappelluti MA et al. 2024. Durable and efficient gene silencing in vivo by hit-and-run epigenome editing. Nature. DOI: 10.1038/s41586-024-07087-8
Cells are biological marvels, but even they need help taking out the trash sometimes.

This is especially true when things go awry!
Cells have mechanisms in place to degrade misbehaving proteins, eliminate things that aren’t needed anymore, or respond to crises by going nuclear.
For cells, the nuclear options are autophagocytosis (self-eating), senescence (cell cycle arrest), apoptosis (programmed cell death), and necrosis (rapid, less programmed, cell death).
But, in some diseases these processes are interrupted and unruly proteins are allowed to continue their delinquent behaviors.
Things can get even worse when cells that should have pushed the red button and sacrificed themselves for the good of everyone else, don’t.
So, how can we use proteomics to get these cells back on track (or obliterate them)?
Well, cells have two distinct protein waste removal services that we can take advantage of:
The Proteasome - These are like protein wood chippers that float around inside a cell. They’re used to chop up useless, misfolded, damaged, or no longer needed proteins!
The Lysosome - Is a specialized membrane enclosed organelle (kind of like an industrial garbage dump) where bulk protein degradation activities take place.
Targeted protein degradation using these two pathways has been an important area of therapeutic development in the last few years.
This work has focused on the creation of ‘chimeric’ molecules that combine the function of two or more molecules together to perform a desired function:
PROteolysis TArgeting Chimeras (PROTACs) - The proteasome degradation pathway is activated when the proteasome comes across proteins that have been tagged with a molecule called ‘ubiquitin.’ This tag is added to proteins by an enzyme, E3 ubiquitin ligase. So, most PROTACs are composed of two components, one that binds the protein we’d like to degrade, and another that binds E3! This brings that protein into very close proximity to the enzyme that marks things for destruction - bada bing, bada boom - no more protein!
LYsosomal TArgeting Chimeras (LYTACs) - The lysosome degrades lots of things including food that’s been brought in from outside the cell (phagosomes), receptors that once resided on the cellular membrane (endosomes), and larger cellular structures like mitochondria (autophagosomes). How all of those things get to the lysosome is pretty well known, and chimeric molecules can be engineered to cause target proteins to be scooped up in one of those ‘-somes’ and degraded in the lysosome! Just like PROTACs, these chimeric combinations involve binding to a target of interest on one end, and to a lysosomal targeting protein/molecule on the other!
PROTACs and LYTACs have shown promise as adept therapeutic garbage men in inflammatory disease (degrade misbehaving receptors), neurodegeneration (target plaque forming proteins), and cancer (initiate cell death).
See, taking out the trash isn't so boring after all.
One of the most important papers in the history of genetics was basically ignored when it was published in 1944.
But, before we get to THAT paper, we need to go back one paper to set the stage.
In 1928, Fred Griffith published, ‘The Significance of the Pneumococcal Types,’ where-in he identified the ‘rough’ (doesn’t kill mice) and ‘smooth’ (kills mice) types of pneumococcus (a bacterium).
Through a series of experiments he showed that if you inject mice with rough, they live, and with smooth, they die.
If you heat kill smooth and inject that, they live.
These are not surprising results.
But, if you inject mice with both rough and heat killed smooth, they die.
The killed smooth type is able to ‘transform’ the rough type of pneumococcus into the deadly type.
Griffith’s transformation experiments are considered one of the cornerstones of molecular biology, but up until 1944, basically the entire field of biology thought the ‘transforming factor,’ or genetic material, was protein.
So, suffice it to say that when Oswald Avery, Colin Macleod and Maclyn McCarty published their paper in 1944 showing unequivocally that DNA was the genetic material, it wasn’t very well received.
Basically no one cared.
Mostly because everyone thought the genetic material was protein and that Avery and team screwed up their experiments.
Except, they didn’t.
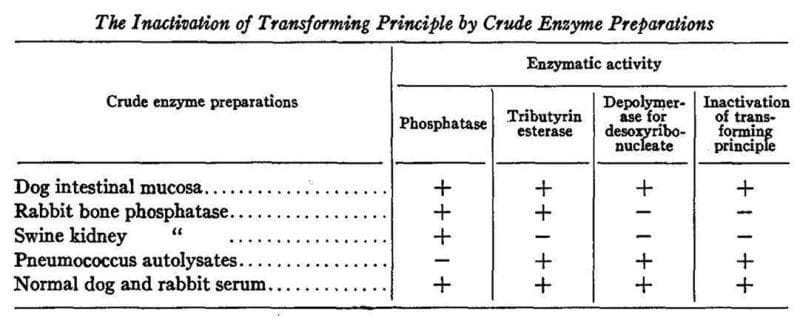
The table above shows an experiment where alcohol purified DNA was exposed to five different crude extracts each with different enzyme activities: phosphatase (degrades RNA, RNase), tributyrin esterase (degrades fats and proteins, protease), depolymerase for desoxyribonucleic acid (degrades DNA, DNase).
In this figure you'll see the last column says 'inactivation of transforming principle' and the only extracts that inactivate the transforming principle are the ones that contain DNase activity.
Purified proteases also failed to inhibit the transforming activity.
But at the time, the genetic material question was deemed to be solved, it was protein, and one of the ring-leaders of the ‘protein is the genetic material’ crew was Linus Pauling.
Pauling is one of the only scientists to receive a Nobel Prize in two different disciplines and is the grandfather of modern structural biology.
He was kind of a big deal.
Pauling himself didn’t believe DNA was the genetic material and instead focused his efforts on studying protein structures.
It was the Hershey-Chase experiment in 1952 that finally convinced him that DNA was the real deal but by that time it was too late.
Watson and Crick, two scientists inspired by Pauling’s work, solved the structure of DNA in 1953 (with help from Rosalind Franklin and others).
But you can't feel too bad for Pauling, he didn't need 3 Nobels anyway.
###
Avery, O et al. 1944. Studies on the Chemical Nature of the Substance Inducing Transformation of Pneumococcal Types. J Exp Med. DOI: 10.1084/jem.79.2.137
Were you forwarded this newsletter?
LOVE IT.
If you liked what you read, consider signing up for your own subscription here:
