Omicly Weekly 2
December 10, 2023
Hey There!
Thanks for joining me for the second BETA issue of Omic.ly!
I'm still trying to work out a few bugs and I appreciate you for bearing with me.
I got a couple of good suggestions since the last newsletter.
One of these was to update the "reply-to" address so it actually goes to me!
And another great suggestion was to add a little table of contents here at the top so you know what you're getting yourself into before you start scrolling.
As always, please send me any comments or suggestions for how I can improve the newsletter.
Please enjoy!
In this week's newsletter you will find:
1) The myth of the $100 genome
2) FFPE is great for histology and terrible for molecular testing
3) The most brazen scientific Easter Egg of all time
The $100 genome is a myth, let's focus on the $0 one instead.

One of the biggest disappointments in the 'cost of a genome' discussion is that despite reducing costs, access to genome sequencing for disease diagnosis is very hard to come by outside of an academic medical center.
First it was the $1,000 genome, then we got a $500 genome on the NovaSeq, now it's ~$200 on the Xplus and Aviti with a sub $100 genome on the DNBSEQ-T20.
The problem is these numbers mean basically nothing to the vast majority of patients waiting to get a diagnosis.
Why?
Well, for one, that number doesn't include the cost of labor, analysis, or interpretation.
In the best case, for a clinical workup of a singleton (the affected individual), we're looking at $50 for sample collection, DNA extraction, library prep, and labor; $100-200 for sequencing; and then between $300 and many thousands of dollars for analysis/interpretation.
If you're doing trios (Mom, Dad, affected individual), you can basically triple those costs.
So even in the best case scenario, we're talking ~ $400, and that's just Cost of Goods Sold (COGS) without adding in any profit.
If you're doing this with a major non-profit, it's ~$9,000 for a trio, but that includes case review by a world class clinical team.
It's likely that equivalent quality can be had for much less, and the future integration of AI and other emerging methods should make these prices more palatable to insurance companies.
And that's what I'm most excited about:
The $0 genome.
Because these $9,000 clinical workups are generating the evidence needed to get insurance companies to pay for them.
My hope is that the majority of payers will finally wake up and say, "Maybe it doesn't make sense for these kids and families to wait a decade for a diagnosis?"
We're starting to see some of this already in more progressive states where they're funding rapid neonatal sequencing programs for very sick kids - California, Michigan, Oregon, Maryland, Minnesota, and others.
But many of these programs have pre-authorization requirements that restrict access to this much needed testing.
Even UnitedHealthcare announced this year that it was going to cover sequencing but only when a genetic cause is suspected and presentation includes any of the following: "multiple congenital anomalies (must affect different organ systems); moderate, severe, or profound intellectual disability diagnosed by 18 years of age; or global developmental delay, or epileptic encephalopathy with onset before 3 years of age."
This is progress but coverage restrictions like these mean a major swath of the population that are affected by late onset genetic disease, and who could benefit from a genetic diagnosis, will be missed.
Unfortunately, there's still work to do, and that includes expanding coverage to carrier, inherited disease, and risk factor screening in healthy people.
Baby steps, I guess.
Formalin-fixed, paraffin-embedded (FFPE) tissues in oncology diagnostics: is there a better way?

But, before we get to that, what is FFPE?
Formalin-fixation was discovered as a cellular preservation technique in the late 19th century and quickly became the most common embalming method throughout the 20th century.
While useful in preserving dead people and animals, it's also quite good at preserving tissues for scientific analysis because it chemically 'cross-links' proteins together making them much harder to degrade.
However, to prevent tissues from shriveling up and losing their morphology, the preservation technique requires a physical stabilizer.
Enter paraffin wax.
After formalin-fixation completes (~24hr), the tissue is embedded in a wax block.
This is followed by 'sectioning' with a microtome (cuts razor thin slices), and finally mounting the tissue sections on a microscope slide for viewing.
This process is great for understanding cellular pathology, but terrible for molecular analysis because getting all of the good bits out of an FFPE sample is very challenging.
Pros:
Preservation of cellular morphology and proteins for imaging studies
Cons:
Everything else
Formalin damages nucleic acids: the fixation process causes DNA and RNA to fragment and this degradation can continue even during cold storage. This can result in a significant reduction in signal, which is important in samples with low tumor fractions.
Fixation can introduce base modifications: deamination of cytosine to uracil and transitions (A>G,T>C) are most common. Base modifications are never ideal because they can result in the reporting of false positive results.
Loss of long range information: because formalin fixation breaks nucleic acid, long range sequence context is lost. Meaning, it's harder to detect gene fusions or chromosomal anomalies using traditional sequencing methods.
Sequence chimeras are prevalent: the damage caused during fixation creates sequence overhangs or single stranded DNA that ends up recombining to create new 'chimeric' sequence that didn't exist in the original sample.
De-paraffinizing a sample is not fun: this involves xylene or intense sample sonication with specialized equipment. One method is unpleasant, the other is expensive.
Treatment with enzymes during processing can reverse some of the damage, but they don't work miracles.
Fortunately, there are alternatives to FFPE and because we know formalin damages nucleic acid, we've been able to develop new methods that are more gentle on the molecular components we're most interested in testing.
These include:
Processing fresh tissue
Flash freezing in liquid nitrogen
Preservation in a tissue stabilizer solution like AllProtect or RNAlater
However, the biggest hurdle is updating the clinical workflow to add the collection of a molecular friendly specimen (in addition to the ones that end up in the jars of formalin!)
This is probably the most brazen scientific easter egg of all time:
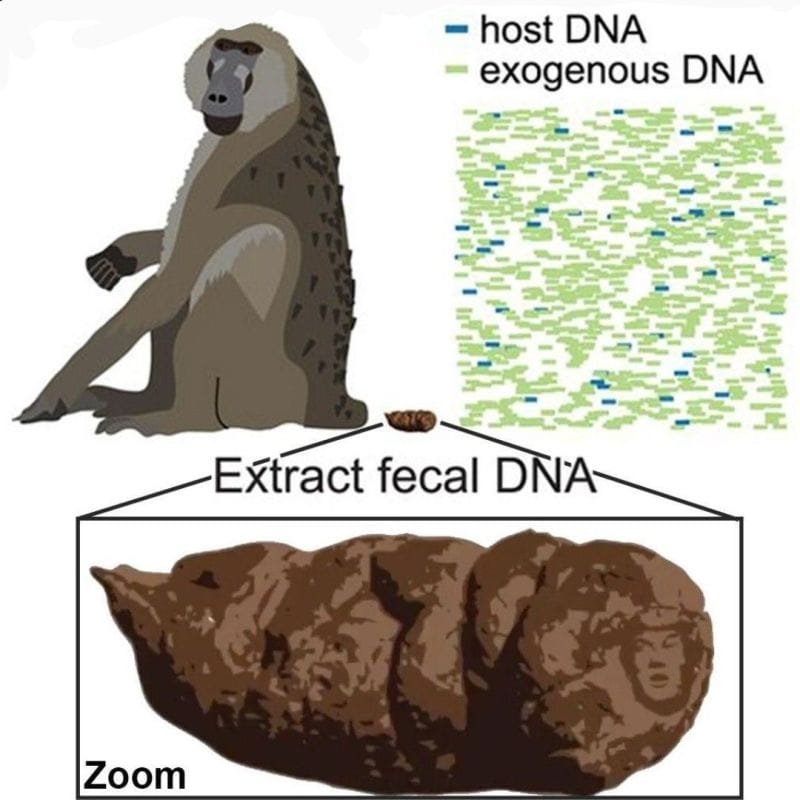
At the end of January 2018, a paper, 'Methylation-based enrichment facilitates low-cost, noninvasive genomic scale sequencing of populations from feces,' was published in Scientific Reports.
Seems innocuous enough, right?
In the article the authors describe sequencing animal populations using non-invasive DNA isolation from feces and they developed a methylation based enrichment method to separate the host DNA from all the stuff in the poop.
It's a pretty ingenious method with applications that extend beyond animal populations.
They even had a fun figure, Figure 1, with their research subject, a baboon, and a little turd to illustrate how their method worked.
The figure is really pretty precious.
But it turned scandalous if you zoomed, like, zoomed a lot.
Because on the tip of the high resolution baboon dropping was a monotone image of Donald J. Trump.
Lil' Donny was discovered shortly after the 2018 election and Nature Publishing Group quickly removed the likeness and provided this sanitized explanation:
"In the original version of this Article, there were unusual aspects to the ‘Extract fecal DNA’ illustration in figure 1. These features have been removed."
But, the internet never forgets 'aspects' that are this 'unusual,' and the doody with the Donald lives on.
###
Chiou KL, Bergey CM. 2018. Methylation-based enrichment facilitates low-cost, noninvasive genomic scale sequencing of populations from feces. Sci Rep. DOI: 10.1038/s41598-018-20427-9
