Omic.ly Premium 55
December 23, 2024
Hey There!
Thanks for spending part of your holiday with Omic.ly!
This Week's Headlines
1) HOT TAKE: The week of Sequence-mas
2) Our skin has it's own distinct immune system
3) The epigenome isn't just some biological afterthought
4) Is the genetic material protein or DNA? A blender decided.
5) Weekly Reading List
The week of Sequence-mas
'Twas the week of Sequence-mas, when all through the industry,
Not a creature was stirring, not even a BALB/c;
The instruments were all running with the utmost of care,
Since we all know Illumina customer service won’t be there;
With fragments nestled all snug in the flowcells,
Visions of nucleotides danced in their nanowells;
And the techs in their labcoats, their eyes all bloodshot,
Staring at clusters and Q-scores and the intensity plot.
When out of the lab arose such a clatter,
I sprang from my desk to see what was the matter.
Away to the bench I flew in a dash,
Hoping no sequencers had been lost in a crash.
The LEDs were casting the eeriest of glows,
Giving a green luster to all of the shadows.
When what to my wondering eyes should appear,
But a flashing red light—oh no! Not this year!
With a silent prayer and an acknowledgement so quick,
I crossed my fingers, hoping it would do the trick.
“You won’t win this time,” I yelled with a grin!
The system responded, and hummed with a din.
As cycles repeated, I smiled with glee,
For the flowcell was flawless, as perfect as could be.
No clogging, no bubbles, no failures this run,
The sequencing concerto, a symphony had spun.
More rapid than reagents, the analysis came,
As I whistled and muttered and called them by name:
"Now FASTQ! Now BAM! Now, VCF and Assembler!
On MiniMap! On PLINK! On, genome browser!
To the end of the reads! To the Telomere wall!
Now map away! Align away! Process it all!"
The reads streamed in, all bright and clean,
Each passing base with a glittering sheen.
And then, in a twinkling, I heard from the cloud,
The results had been processed—I laughed out loud.
As I uploaded the files and refreshed with great care,
The final data appeared with metrics to spare.
The variants were called, the sequences aligned,
The output so glorious, it blew my mind.
Exomes and genomes, bases complete,
A holiday miracle—the dataset was sweet.
With a sigh of relief, I strode out of sight,
"Happy Sequence-mas to all, and to all, a good night!"
###
We'll get back to the goss next week.
I hope you all have a happy holiday!
Our immune system plays a critical role in recognizing, fighting, and preventing infections, but now we’ve discovered that our skin has its own!
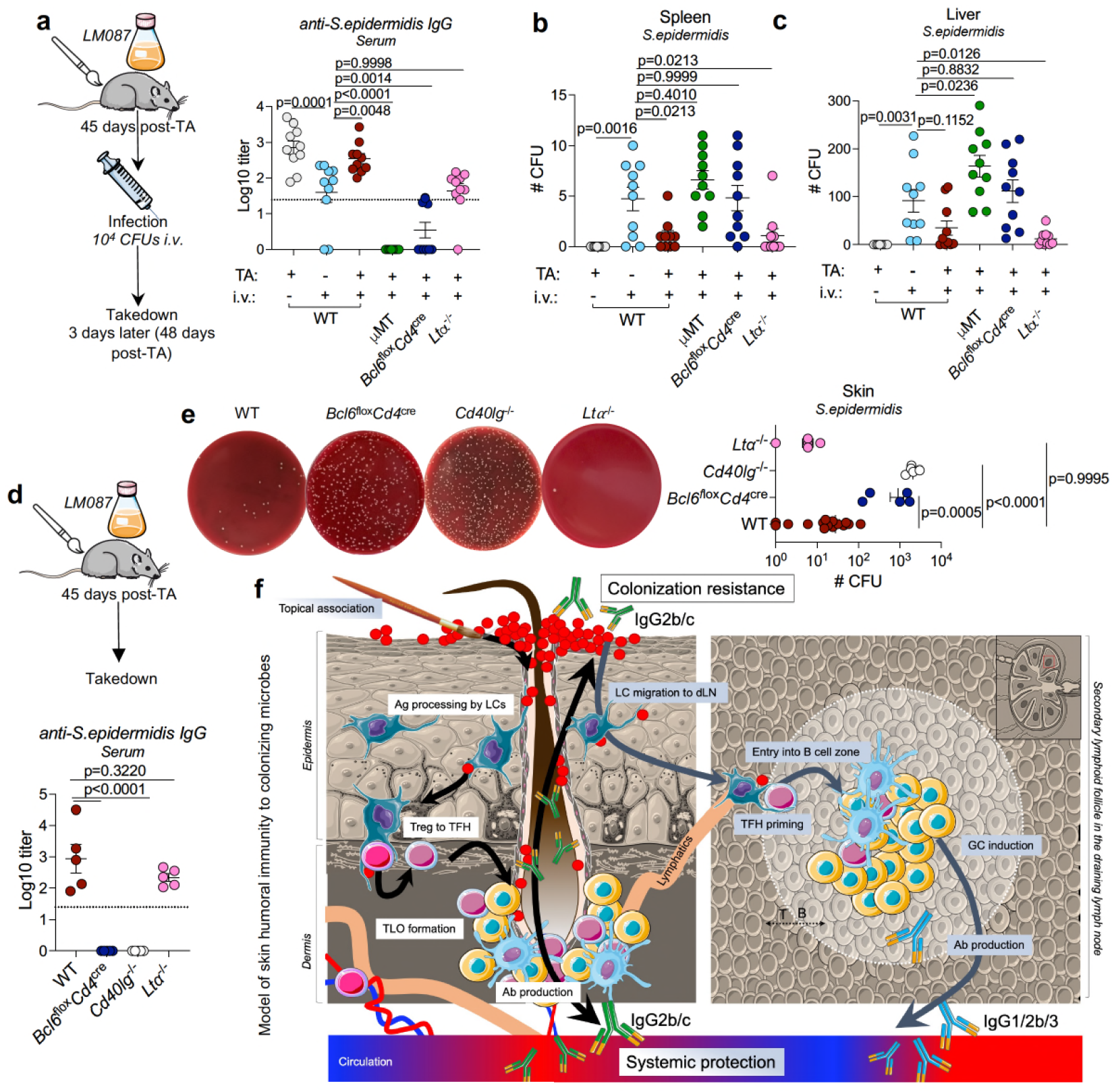
Our skin and mucosa are our first line of defense against infection with deadly pathogens.
But these surfaces and membranes aren’t sterile environments!
They’re teeming with biological activity and because they provide this “barrier” function, they’re constantly fending off infection from pathogenic microbiota.
And we call this a barrier, because that’s exactly what it is!
Our mucosa are like a moat around a castle that slows down any invaders (or drowns them if they can’t swim!)
And our skin is like the castle wall, helping to physically prevent big and small things from entering our bodies.
But that’s not where our skins’ defenses stop!
We’ve known for some time that the immune system is active in the skin and that it plays a vital role in managing the communities of microbes that live in and around our bodies.
This is important, because not all of the microbiota that we encounter are bad!
So our immune system has to play this tricky dance of letting the good guys in and keeping the bad guys out.
We now know that it does this using its own localized immune regulatory system!
In this week’s paper, researchers showed in mice that the skin has its own localized immune system that acts (mostly) independently.
In the figure above they a) topically associated (paint brush) a commensal (good) bacteria, S. epidermis with wild-type mice (WT), Bcl6Flox/cd4cre mice that can’t make B cells (make antibodies), µMT mice that can’t make T Helper cells (stimulate b cells to make antibodies), and Ltα-/- mice that lack secondary lymph nodes. Mice were then injected with S. epidermis 45 days later to see their immune response (a,b,c graphs). WT and mice lacking secondary lymphnodes mounted a strong response while mice who couldn’t make antibodies did not! d) is a control showing IgG production was the result of TA and not infusion e) shows representative bacterial growth from the serum of infused mice and f) is an overview of how the skin immune system works!
In this scheme, Langerhan’s cells (LCs) see antigens that collect in the hair follicle. This activates t-helper cells (Treg to TFH) that form complexes with b cells to form a germinal center like structure (where b cells grow up to make and secrete antibodies). These antibodies are the secreted into the follicle to protect the skin and into systemic distribution to the rest of the body.
So, the skin isn’t just some simple barrier!
It is a major part of our lymphatic system helping to make, distribute, and protect the entire body against infection!
In a second paper, the team modified S. epidermidis to display a piece of the tetanus toxin to the skin’s immune system and showed that this could be used to vaccinate mice against tetanus infection.
The implication here (if it all works the same in humans) is pretty mind blowing because it means that needle free vaccines could be closer than many of us ever imagined!
###
Gribonika I, et al. 2024. Skin autonomous antibody production regulates host-microbiota interactions. Nature. DOI: 10.1038/s41586-024-08376-y
The Epigenome: What it can tell us and why we should care to look at it thoughtfully.

Epigenetics is the study of modifications that aren’t coded directly in the sequence of our DNA.
These modifications can include methylation of cytosine DNA bases (they’re still cytosines, though) and modifications to the histones that coil DNA up into chromosomes.
The major effect of these changes is that they result in the opening or closing of our DNA for reading by the cellular machinery that converts this code into proteins.
The epigenome differs from the genome in that these modifications can change frequently and are influenced by a variety of external environmental factors and cellular microenvironmental factors.
Some epigenetic modifications can even be inherited from your parents!
However, it’s early days.
And it’s safe to say that we have A LOT to learn about the epigenome.
There are still many questions about whether epigenetic modifications are drivers (causes) or passengers (effects).
But we do have a lot of evidence that epigenetic changes are important in key areas of human health and disease:
Cancer - Mutations are frequently observed in DNA and histone modifying enzymes, histones themselves, and other genes involved in changing the 3D structure of the genome. Under certain circumstances, it also seems that epigenetic changes can be major drivers of cancer progression in the absence of other obvious changes to DNA. All of these combined serve to turn on tumor promoting genes (oncogenes) and turn off tumor suppressors, ultimately leading to cancer!
Aging - Often referred to as the epigenetic clock, it has been observed that the amount of methylation in the genome changes as we age! These changes have been correlated with cellular dysfunction (loss of control, cell death, etc) and are exacerbated by obesity and low levels of physical activity.
Rare Diseases - Mutations in epigenetic regulatory genes can cause a number of rare diseases. These are often characterized as traditional single gene diseases, but they underscore how important it is to have a properly functioning epigenome. There is also quite a bit of research on the role of epigenetics in rare complex diseases like Schizophrenia and Autism Spectrum Disorder where it has been notoriously hard to pin down major genetic causes.
Disease Risk - Exposure to environmental factors such as drugs, chemicals, infections, or other stressors in the early stages of development or early years of life can cause lasting epigenetic changes that may result in disease later in life. These can include diabetes, obesity, cancer and neurological disorders. But one important new area of research is how behavioral stresses in adolescence can contribute to epigenetic changes that lead to the development of psychiatric disorders.
That said, there’s no doubt we should be excited about the future of epigenetics and all of the secrets we still have to learn from the epigenome!
Deoxyribonucleic acid (DNA) is the genetic material, but early on, most of science thought the genetic material was protein. That changed in 1952.
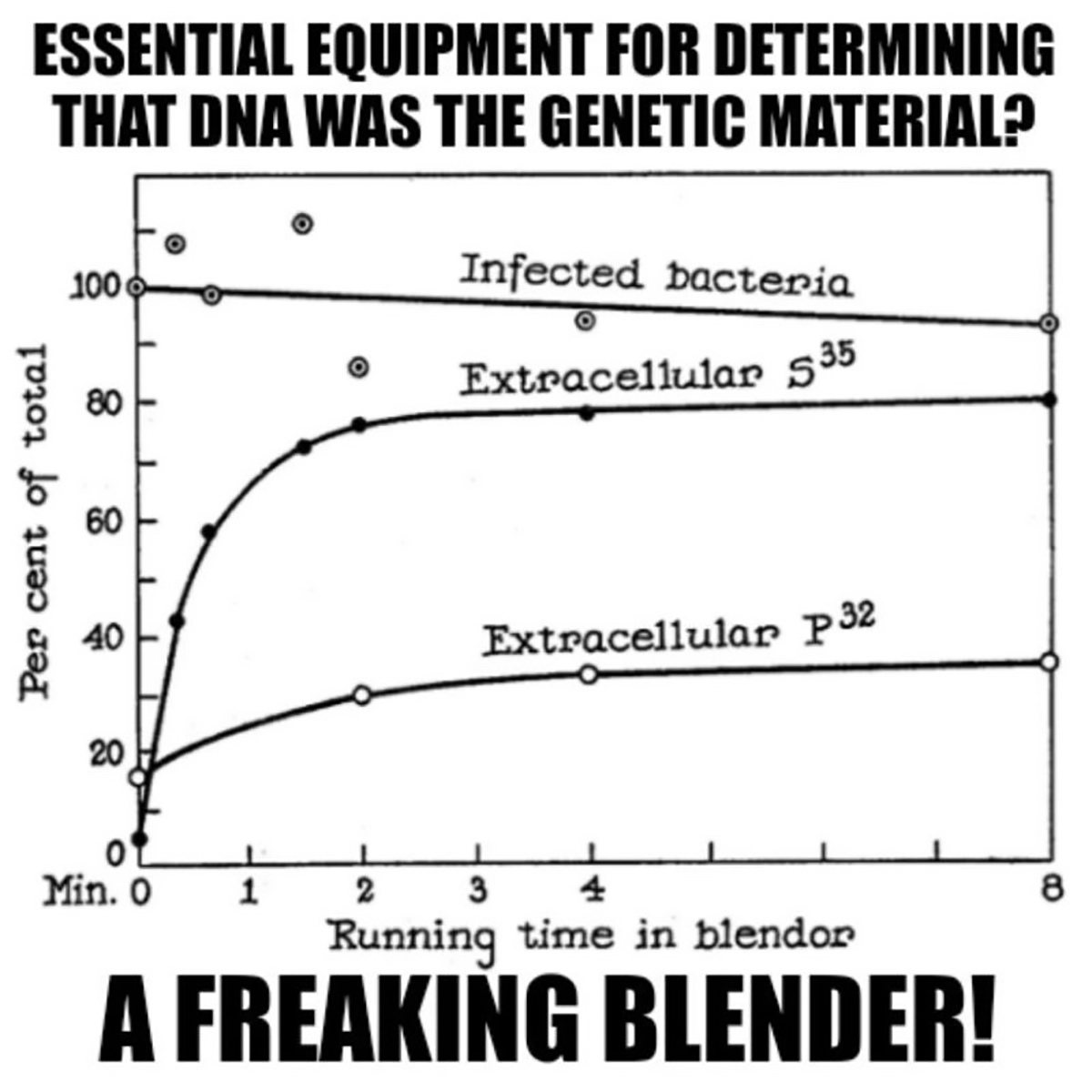
Ok, it didn’t change overnight, because scientists are pretty stubborn.
It actually wasn’t accepted until the 60’s, but, in 1951, Alfred Hershey and Martha Chase began a series of experiments that conclusively showed that DNA was the genetic material.
There was already good evidence that the genetic material wasn’t protein, but early molecular biologists couldn’t wrap their heads around how 4 bases of DNA could code for all of the diversity we see, especially when proteins are made up of long chains of 20 amino acids.
Surely, since 20 is greater than 4, you get more diversity with 20!
So the hypothesis was that DNA must be the scaffold that holds everything together.
Of course, we know now that’s absolutely not true, and it’s basically the opposite - DNA is the genetic material and protein serves a support function.
The work that synthesized all of these ideas together was completed by Hershey and Chase in what are now referred to as the Hershey-Chase Experiments.
They worked with bacterial viruses called bacteriophages.
These are very simple viruses that consist of a protein shell surrounding the DNA of the virus.
But before the 1950’s, no one knew what phages looked like or how they infected bacteria.
In 1951, Thomas F. Anderson, a pioneer of electron microscopy, showed that phages are like little moon landers and attach themselves to the outside of the bacteria but do not enter them.
This created a bit of a conundrum, how can protein be the genetic material if none of the phage protein gets inside?
Generating the figure ainvolved Hershey-Chase, a Waring blender and phages grown in the presence of radioactive food items.
Luckily, proteins do not contain Phosphorus (P) and DNA does not contain Sulfur (S), so radioactive P-32 and S-35 could be used to label each component of the bacteriophage individually.
Radioactive phages were then combined with bacteria and stuck in a blender.
Hershey and Chase discovered that this technique didn’t kill the bacteria, but it was perfect for shaking the empty phage husks from the bacteria.
This is documented in the figure which shows 90% viability of the infected bacteria but that 80% of the protein (S35) remained external while only 30% of the DNA (P32) does after blending.
This result supported the electron microscopy observation that the phages are basically just little protein covered DNA injectors!
Hershey and Chase followed up by showing that 30% of the DNA that makes it into baby phages after the blender experiment was radiolabeled while <1% of the protein was, essentially proving that DNA, and not protein, is the heritable genetic material.
###
Hershey, AD and Chase, M. 1952. Independent Functions of Viral Protein and Nucleic Acid in Growth of Bacteriophage. J Gen Physiol. DOI: 10.1085/jgp.36.1.39
Weekly Reading List













Were you forwarded this newsletter?
LOVE IT.
If you liked what you read, consider signing up for your own subscription here:
















