MRD Testing: It's just a phase
All of the variants are getting in on MRD testing, even the phased ones!
Minimal Residual Disease (MRD), also known as Molecular (when looking at nucleic acid) and Measurable (because, why not make this confusing?) Residual Disease describes the amount of cancer left in the body after treatment.
Measuring MRD over time allows us to know when a cancer might be coming back.
It also can help us make decisions about what treatments to use, or tell us if an adjuvant therapy (treatment, like chemo, given after a tumor is removed) is working.
But determining residual disease in patients has historically been pretty challenging.
And, up until the invention of massively parallel high throughput sequencing, knowing when or if a cancer might recur was almost impossible.
That’s changed now that the price of sequencing has come down drastically in the last two decades.
It’s now cheap enough to use routinely to help us find early clues about cancer’s recurrence.
We can do this because normal cells and cancers dump some of their DNA into the bloodstream as they grow.
These DNA fragments can be sampled in a blood draw and the “cell free” or “circulating tumor” DNA (cfDNA or ctDNA) can be sequenced to determine if the fragments are regular old normal DNA or DNA that came from a cancer.
This is usually done by looking for single nucleotide variants (SNVs) and comparing those variants to a person’s healthy normal tissue.
Fragments found to have differences could indicate that a cancer is present.
Or they could just be random errors that get introduced during the process of manipulating a sample.
These errors, unfortunately, can sometimes end up producing a false-positive result.
So, we inevitably end up sequencing a bunch of DNA to try find all of the needles (ctDNA) in the haystack of DNA that’s circulating in our blood (cfDNA) to be really really sure we haven’t made any mistakes.
But wouldn’t it be nice if we had a better way to determine which fragments of DNA came from tumors, and which were just the product of weird errors made by the enzymes that we use to process these samples?
We’ve invented a couple of interesting solutions here that involve clever barcoding schemes, but these can add significant cost to the process.
A group of researchers recently wondered if they could do this more simply using phased variants to both reduce the false positive rate and track tumor DNA within the bloodstream.
Phased variants are just variants that occur on the same DNA fragment.
And because cancers mutate frequently, they often have more phased variants in their genomes than are found in healthy tissue.
With this in mind, the researchers behind today’s paper developed a method called Phased Variant Enrichment and Detection Sequencing (PhasED-Seq) and validated its use as an MRD test in large B-cell lymphoma (DLBCL).
Here, the test starts by sequencing tumor and healthy DNA to develop a tumor specific phased variant (PV) list.
That list is then used for subsequent monitoring of plasma samples to see if those same PVs reappear in the bloodstream!
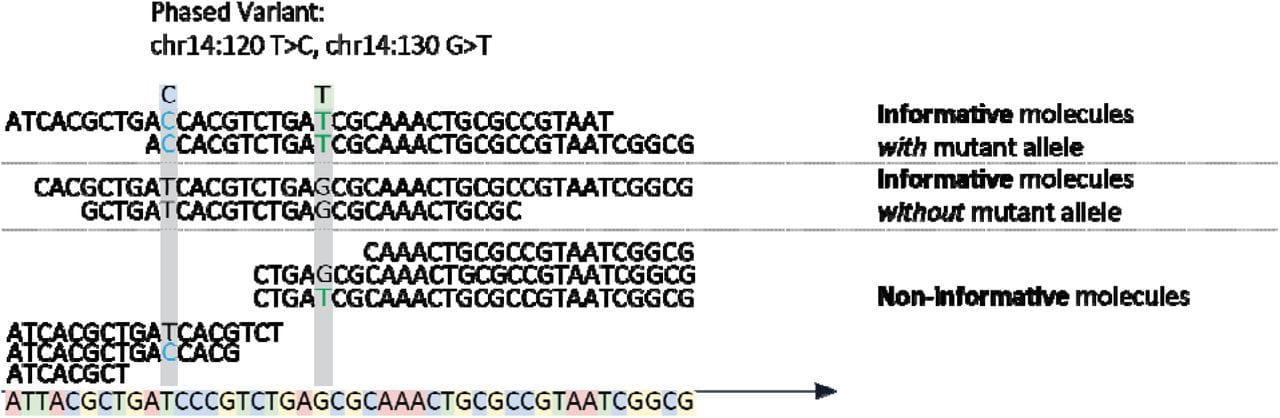
The figure above shows the basics behind the method which involves the capture of DNA fragments from regions known to be frequently mutated in DLBCL followed by short-read high throughput sequencing to detect PVs.
Informative molecules are those that span two phased variants, uninformative ones are those that span the region but only contain one or none of the variants.
In their analytical validation, they show that their test using PVs had a very low false positive rate (0.24%), a limit of detection of 0.7 parts in 1,000,000 and a precision of more than 96%.
That’s pretty good!
And with so many of these high performing MRD tests coming to market, it will be exciting to see how these new tests will help to extend the lives of cancer patients.
