A giant, million sample genetic database was released in November
How gigantic?
Well, let's first put it into perspective with the other available databases:
UKBiobank: ~500,000
GnomAD: ~125,000
ExAC: ~60,000
EVS: ~6,500
1KG: ~2,500
The Regeneron Genetics Center Million Exome (RGC-ME) set is the largest of them all and includes 985,830 samples!

It more than doubles the number of samples we previously had for rare variant and population studies.
But before we get too excited, it's important to explain what an Exome is and how it differs from a genome.
The human genome is 3 billion base pairs spread across 23 chromosomes, but only about 65 million of those base pairs, or ~2%, code for protein.
This portion of the genome is referred to as the Exome and about 85% of disease causing variation occurs in the exome so while it's not all 3 billion base pairs, it covers most of the variation we care about from a genetic disease perspective.
From a broad research perspective, genomes would be way better, but a massive exome dataset like this is a goldmine for a pharmaceutical company like Regeneron who is looking for variants with interesting phenotypes that indicate potential pathways to target for therapies.
One recent example here is the discovery of the gene PCSK9 as a therapeutic target for the reduction of cholesterol. It was discovered that human 'knockouts' for PCSK9 were correlated with exceptionally low cholesterol and a potential blockbuster drug was born.
So, a million sample database can be used to find similarly useful therapeutic targets but much like our other genetic databases, this one also isn't very diverse.
For context, the demographics of the United States are:
White: 58%
Hispanic: 19%
African: 12%
Asian: 6%
Other Ethnicities: 5%
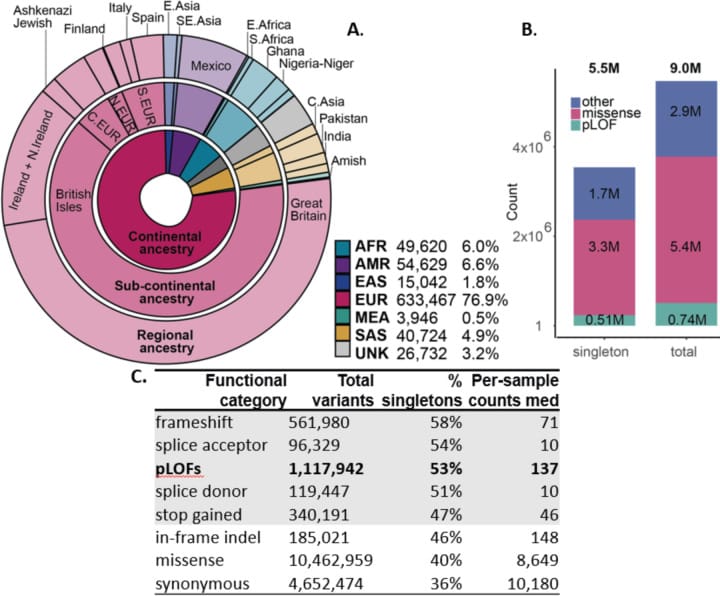
This data set is 77% white, 6% African, 5% Asian and 7% admixed American.
Despite the skew, it's still the most diverse dataset in existence just because of its sheer size.
And that means it can be used to help clean up some of our blindspots!
The researchers discovered that fewer pathogenic variants were found in non-whites but that non-whites also had more variants of unknown significance with Africans having the greatest number of variants in their genomes when compared to the standard reference.
These results highlight the need for both reference diversity and including diverse populations in studies so that pathogenic variation can be identified across all ethnicities.
To this end, RGC-ME was released in aggregate as a part of a public variant browser similar to ExAC/GnomAD with fine breakdowns of variant allele frequencies by population.
This is huge because RGC-ME includes, "10.5 million missense (54% novel) and 1.1 million predicted loss-of-function variants (65% novel, 53% observed only once)."
