So, you want to learn more about your microbiome?
Cool, here's what you need to know!
Cool, here's what you need to know!
The human microbiome is the entire community of microorganisms that live on or in your body and is made up of bacteria, fungi and viruses.
Your body contains, or has on it, many different microbiomes.
This is because each crack, crevice, surface, and orifice creates a distinct environmental niche that promotes the growth of totally different microorganisms!
Meaning, your skin, your mouth, your armpits, and your gut have different microbiomes.
But it doesn't stop there, your gut microbiome itself can be segmented into different microbiomes because your upper and lower gastrointestinal tract create completely different environments.
So If you want to learn about your microbiome, you need to pick one, or choose a service that gives you multiple swabs to test them all!
Well, all except your upper GI, that one's a bit tough to do yourself, even if you have a reeeeallllly long swab.
Once you've settled on which one you want to test you need to pick a service provider.
This is a pretty crowded space but I'm going to let you in on a little secret.
Most of these tests are actually just a resold product from a handful of vendors who provide 'white label' services, so you're mostly getting the same thing from everyone.
And what you're generally getting is a read-out of your microbial community based on 16s ribosomal sequencing.
Now, only bacteria contain a 16s gene so the vast majority of services aren't really reporting out a true microbiome because they're missing all the fungi and viruses!
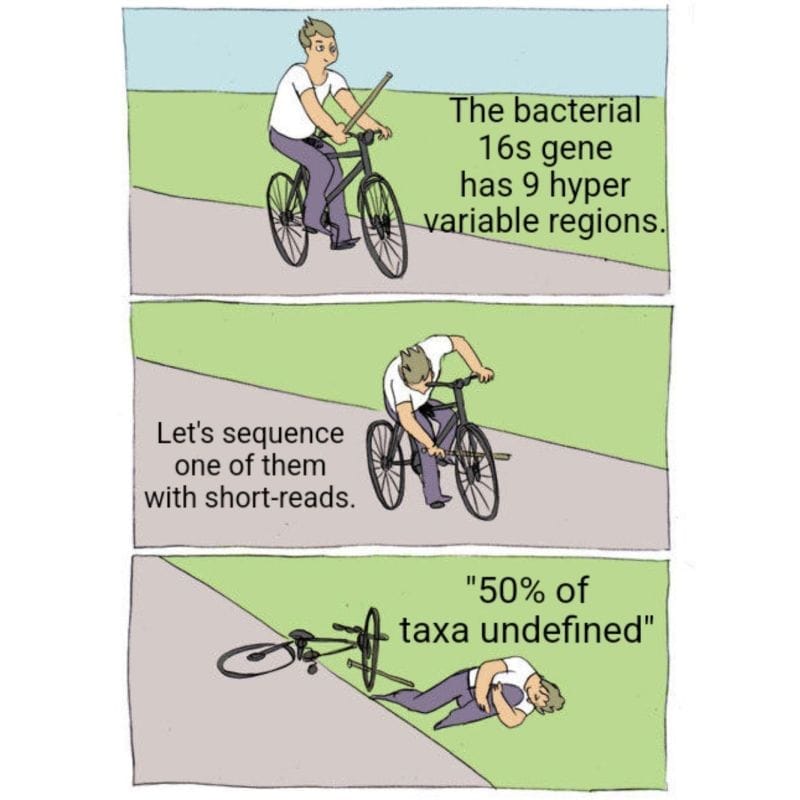
But to make matters worse, most of these services only sequence a small fraction of the 16s gene, the V4 hypervariable region.
You might be thinking, 'hypervariable sounds good!'
Except the 16s gene is 1600 bases long and has 9 hypervariable regions...
Sequencing just one of them is not going to give you the most accurate representation of your community.
And if you really want to do this right, you'd do 16s-ITS-23s.
Except you can't sequence that unless you use **drum roll** long-reads because it's ~4,500bp!
Microbiome sequencing is yet another example of a situation where 'friends don't let friends short-read,' especially if you actually care about identifying species or subspecies level community composition.
There are products that claim they can get species level descrimination from just V4.
But don't be fooled!
While they can detect about 50% of species with just V4, you really need all 9 hypervariable regions to get a species level identification for all of the taxa in a sample.
Otherwise, the best you can do is a genus level identification.
So, what do you actually get out of doing a direct to consumer microbiome analysis?
That's a good question.
Because there's general agreement that the microbiome is important, but what we can actually learn by sequencing it, other than saying there's some bacteria there, is still a work in progress.
