Metagenomics hits the clinic to diagnose CNS infections
Whole genome metagenomics is better than the standard of care in detecting the causal bugs of CNS infections
Unfortunately, whole genome metagenomics has struggled to gain a foothold in the clinic.
This is largely because this testing is expensive, and at least in the US, insurance companies refuse to pay for things where there isn’t sufficient clinical evidence to justify those costs.
At least that’s been the excuse they highlight in their refusals to pay for such innovative and life savings tests.
However, new studies, like the one shown in the figure above, will hopefully start reversing those coverage denials.
In diagnostic testing, speed to result is always of critical importance and we often optimize processes to be sure that samples are received and processed in the shortest “turn around time” possible.
This is because every result is a patient, and many of the tests that we perform, especially the ones that diagnose infectious diseases, are used to help prescribe medications.
And in the case of infections, prescribing the right medication at the right time can mean the difference between life or death.
This is a notable problem in infections of the central nervous system (CNS) which can be uniquely challenging.
You’ve most certainly heard of meningitis (inflammation of the membranes around the brain and spinal cord), but there’s also encephalitis (inflammation of the brain), and very rarely there are infections that can cause both of those at the same time: meningoencephalitis!
Now, diagnosing these kinds of infections can be tricky but the standard of care for testing here is serology (use antibodies to detect infectious agents) or direct detection of pathogenic nucleic acids using something like PCR.
The challenge with these types of assays is that you kind of need to know what you’re looking for, and even running a large panel of serology tests or PCR tests can still miss what’s causing the problem!
But we now have high throughput sequencing that can just sequence everything without a doctor having to guess if an infection is viral, bacterial or the result of a parasite!
In this week’s paper, the authors showed 7 years worth of data performing a metagenomic sequencing test (mNGS) for CNS infections and compared its performance to serology and direct detection with other molecular assays.
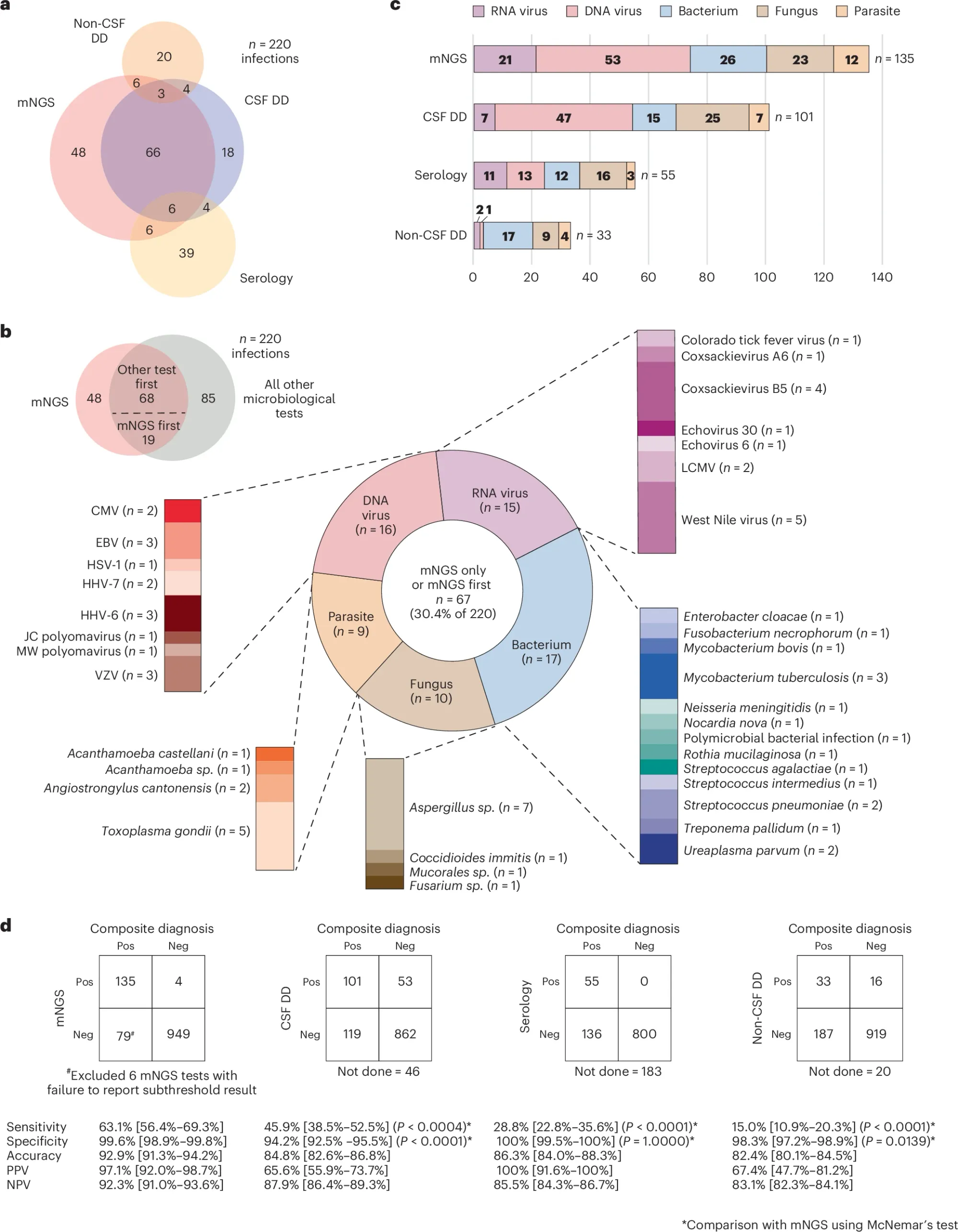
In the figure above a) shows a Venn diagram of all of the infections diagnosed by each technique b) is a breakdown of everything detected by mNGS c) is a bar chart showing all of the different types of bugs that were detected and d) compares the performance of each test to the composite diagnosis (consensus) and highlights that mNGS outperforms all other testing modalities
However, mNGS wasn’t perfect and had false positives and false negatives which the authors dig into and highlight that in many of those cases where there were false negatives that host background was high.
One of the biggest challenges in doing metagenomics is separating the host DNA from the infectious signal.
Here the authors used antibodies to remove methylated human DNA from the samples before processing them, but that process isn’t perfect (and not ALL human DNA can be removed this way!)
So, having too much human DNA around can limit your ability to detect the rarer sequences from the infectious agents!
But at the end of the day, it all comes down to performance and mNGS beat out both serology and direct detection testing which makes a strong case for its use diagnosing critical CNS infections in the clinic.
