Listen up! Fish gills are more important than you might think
Your ears evolved from fish gills!
One of the defining characteristics of mammals are their prominent external ears.
Dogs have them.
Cats have them.
Bats have them.
You have them.
But up until now we didn’t really have any idea how mammalian ears came into being.
Physical structures usually evolve from other things - like fish fins turning into mammalian arms over eons.
However, since ears are made of non-bony cartilage, it’s been impossible to see where external ears came from in the fossil record!
There’s been some speculation that much of the programming for mammalian ears was derived from fish and that fish spiracles (help some fish breathe and are situated behind their eyes) provided the instructions for our inner ears.
But how our external ears sprung forth from our heads, and what fish parts might have aided in that outgrowth has remained a mystery.
That all changed last week with the publication of a paper showing that fish gill cartilage expresses the same genes as human outer ear cartilage.
You might be thinking that this was as simple as isolating ear and gill tissue and seeing which genes each expressed the most.
But gene expression is tricky, and sometimes the most expressed things in a tissue aren’t the most important for their structural development!
Much of the time, particularly when it comes to developmental biology, it’s transcriptional regulation of what gets expressed (and when) that drives the structural development of a tissue.
And with respect to mammalian ears, it looks like we co-opted a fish gill transcriptional regulatory network to do it!
To figure this out, the researchers behind today’s paper decided to look not just at what genes were expressed in ears and gills, but also how that expression was regulated throughout development.
They did this using single-cell RNAseq (gene expression) and ATACseq (chromatin accessibility - ie finds enhancers and active regions) to compare differentially accessible regions (DARs) in the genome.
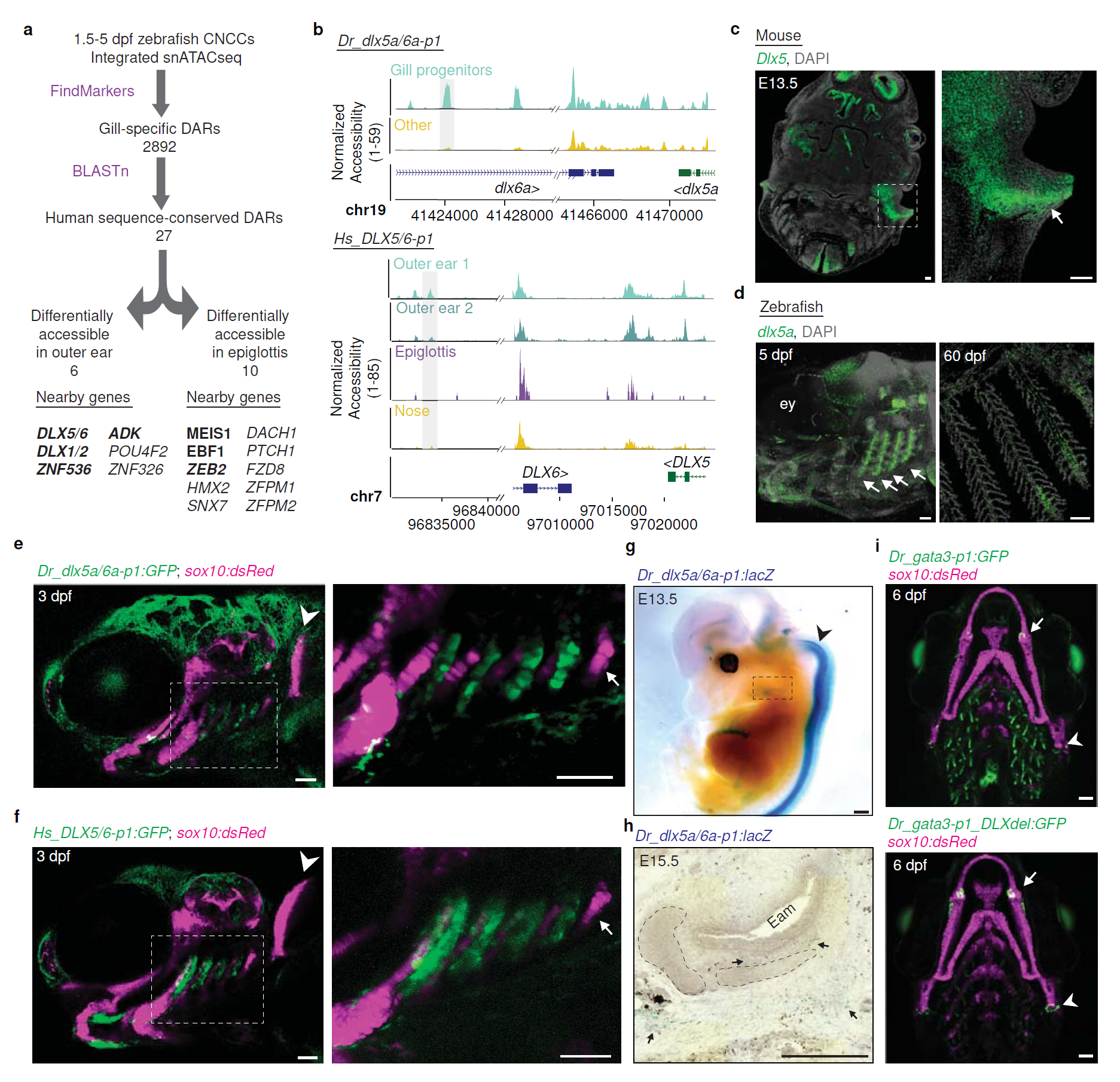
What they found can be seen in the figure above a) schematic of how they identified regions and genes of interest in fish gills and mammalian outer ears - some of the top hits were developmentally important transcription factors (bind to DNA and help turn it into RNA) like DLX1,2,5,and 6
b) DLX5/6 genome accessibility plots comparing fish (top) to humans (bottom) and show that the gill and ear accessibility are quite similar
c, d) RNA in situ hybridization (shows you where the RNA is) of DLX5 in the mouse head (arrow points to the developing outer ear) (c) and fish gills (arrows point to developing gills) (d)
e,f) Human DLX5/6 (f) is expressed in the same places as fish DLX5a/6a (e) when its put into a fish embryo
g,h) and the same is true when you put fish DLX5a/6a into a mouse!
i) is a control showing that when you delete the DLX5/6 DAR, you lose gill expression (green).
Interestingly, mice who have deletions of DLX5 and 6 also lack outer ears, but this is the first time that this gill-ear transcriptional connection has been made!
