Let the Nanopore Wars begin: BGI has a new nanopore sequencer
The Nanopore Wars are coming and BGI just fired the first salvo in what is sure to be an exciting conflict
Nanopores as sequencers have been a genomics dream since at least the late 1980s.
But, the first patent for this style of sequencer was filed in 2008 by Mark Akeson, David Deamer, William Dunbar, Noah Wilson, and Kathy Lieberman at UCSD.
Deamer had originally thought of a ‘channel’ based DNA sequencer that determined the sequence of bases based on how they disrupted an electrical current during a road trip in 1989 and scribbled his thoughts in a notebook.
These notes would serve as the basis for this patent 20 years later.
The journey to that filing obviously took some thought and experimentation.
A meeting between Deamer and his old friend, Daniel Branton, in 1991 reignited his interest in the topic of channel, or pore, based sequencing and the next 17 years were dedicated to the pursuit of figuring out how ‘nanopores’ could be used to make a better DNA sequencer.
The first fruits of this labor were published in PNAS in 1996 where the team was able to show they could pass DNA strands through pores and generate signals.
But it wasn’t until the early 2000’s that the team was able to actually discern nucleotides within those strands.
However, this was good enough evidence that there was potential in nanopores as sequencers and in 2007, Oxford Nanopore Technologies licensed this technology for commercialization.
The rest, as it were, was history.
That might be a little too flippant, though, since significant development effort has gone into converting this proof-of-concept technology into the current product that ONT sells today.
Fortunately (or unfortunately for ONT), since commercialization to a viable sequencing technology has taken over a decade and a half, that means the original patent is set to expire in less than 5 years.
Which is what makes this recent preprint from BGI on their internally developed nanopore technology so interesting.
In it they detail how they have discovered or engineered pores and motor proteins with less than 50% sequence identity to other nanopore proteins.
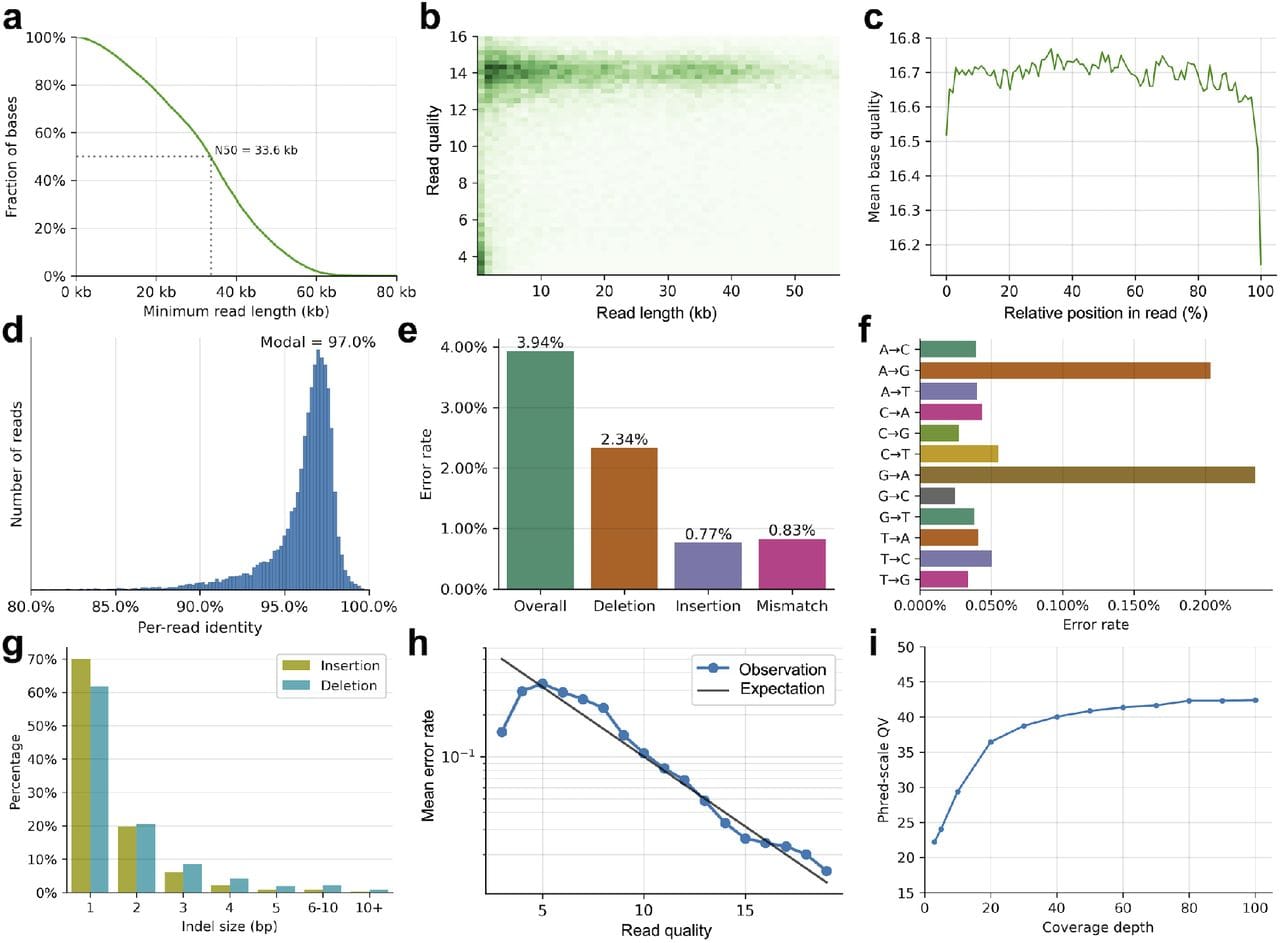
They’ve also packaged this into something that looks and performs similarly to what we’ve all come to expect from nanopores which can be seen in the figure above.
In A) they achieve a read length N50 of 33.6kb (ONT is ~25kb), B,C,D) have a read quality of Q16.7 or ~97% (ONT is Q20+ or >99%), E) Their overall error is dominated by deletions, F) They struggle most with A<>G transitions, and I) their consensus accuracy is on par with currently accepted standards ie >Q30 @ 30x coverage.
Whether this platform struggles with homopolymers (extended runs of the same base) remains to be seen since this is not mentioned anywhere in the paper.
But these results are promising and potentially offer a serious alternative to ONT in the Nanopore sequencing space.
And since they seem to have figured out how to divorce the detection electronics from the pores, they could also offer a significant cost advantage over the competition.
That is if you can wait until 2029 to purchase them in North America.
